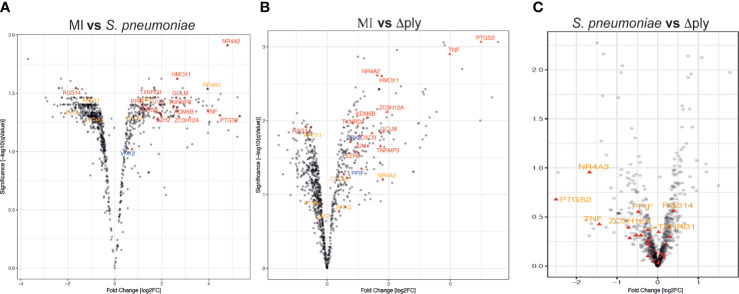
Figure 1.
Oxidative stress response pathway terms significantly upregulated. Monocyte derived macrophages (MDMs) were challenged with either S. pneumoniae, the isogenic pneumolysin negative mutant (Δply) or mock infected with phosphate buffered saline (MI) in biological replicates of three. After 3 h incubation the gene expression was measured using Affymetrix arrays. (A) volcano plots comparing log2 (fold change) to log10 p value for MI vs S. pneumoniae challenged MDMs. (B) volcano plots comparing log2 (fold change) to log10 p value for MI vs Δply challenged MDMs. (C) volcano plots comparing log2 (fold change) to log10 p value for S. pneumoniae vs Δply challenged MDMs. The highlighted genes belong to the Oxidative stress response Gene Ontology term and are significantly expressed (q value <0.05). In red are those found in challenge with both strains, in blue the genes that are only found in the response to Δply challenge and in orange the pneumolysin-dependent genes found after S. pneumoniae but not Δply.