Fig. 2.
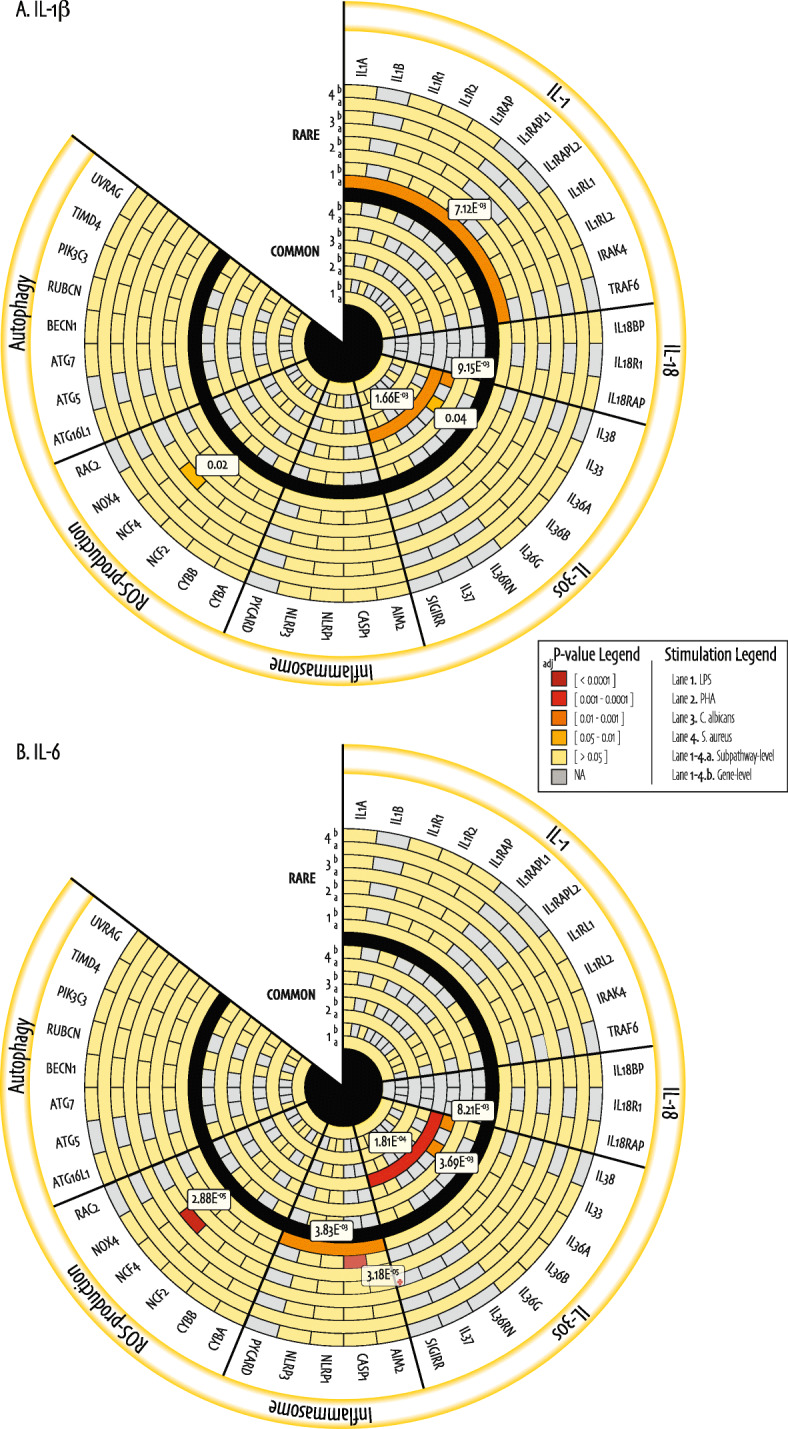
Association landscapes of SKAT (Bonferroni-adjusted) adjP values. The circular heatmaps consist of two rings separated by a black lane, where the inner ring shows the SKAT adjP values with only common (AF ≥ 5%) variants (SKAToC), and the outer ring the SKATO adjP values with only rare (AF < 5%) variants on the gene and subpathway levels with log10-transformed IL-1β (a) and IL-6 (b) cytokine production respectively. Each ring consists of 8 lanes that represent different stimuli; (1) LPS, (2) PHA, (3) C. albicans, (4) S. aureus, (a) showing the subpathway-level result and (b) the gene-level result. The gene names at the surface of the outer ring of the heatmap are grouped based on corresponding subpathway as annotated on the yellow border, and genes without identified genetic variants are not shown. Genes or subpathways without identified genetic variants contributing to a particular association (i.e., for a gene with a single rare variant the gene-level output is not considered, but it does contribute to the subpathway-level association) have been assigned the value NA and are shown in light gray. Significance of P values is highlighted in color, and only significant P values are labelled. Annotation: * = the CASP1 set-based association was the only one not confirmed by Spearman correlation. Abbreviations: SKAT = Sequence Kernel Association Test; LPS = Lipopolysaccharide; PHA = Phytohaemagglutinin; C. albicans = Candida albicans; S. aureus = Staphylococcus aureus
