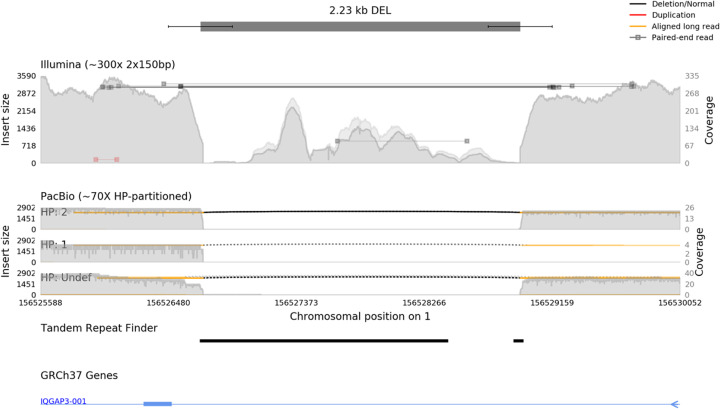
Fig. 1.
Samplot creates multi-technology images specialized for SV call review. A putative deletion call is shown, with the call and confidence intervals at the top of the image (represented by a dark bar and smaller lines). Two sequence alignment tracks follow, containing Illumina paired-end sequencing and Pacific Biosciences (PacBio) long-read sequencing data, each alignment file plotted as a separate track in the image. PacBio data is further divided by haplotype (HP) into subplots. Reads are indicated by horizontal lines and color-coded for alignment type (concordant/discordant insert size, pair order, split alignment, or long read). The coverage for the region is shown with the gray-filled background, which is split into map quality above or below a user-defined threshold (in dark or light gray respectively). An annotation from the Tandem Repeats Finder [16] indicates where genomic repeats occur. A gene annotation track shows the position of introns (thin blue line) and exons (thick blue line) near the variant; a small blue arrow on the right denotes the direction of transcription for the gene