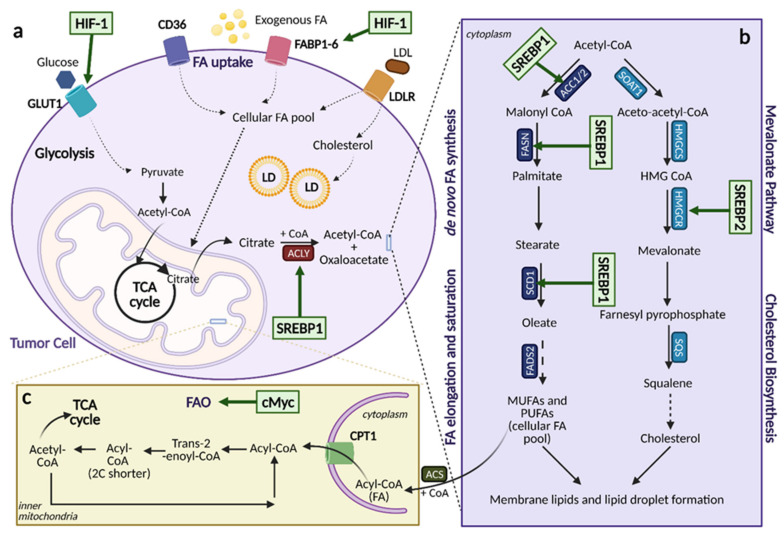
Figure 6.
Lipid metabolic reprogramming in Cancer. An overview of lipid metabolic pathways and how they are modified in cancer. (a). Tumor cells take up fatty acids (FAs) using multiple trans-porters, including CD36, FA binding proteins 1-6 (FABP1-6), and a low-density lipoprotein receptor (LDLR) for low-density lipoproteins (LDL). These free FAs then become a part of the cellular FA pool where they can enter the citric acid (TCA) cycle and contribute to lipid formation. The upregulation of FA uptake in cancer occurs through hypoxia-inducible factor (HIF-1)-induced FABP1-6 over-expression. (b). The upregulation of lipogenesis and cholesterol biosynthesis is achieved through sterol regulatory element binding protein (SREBP) activation. SREBP1 activation induces the ex-pression of lipogenesis genes, while SREBP2 activation induces the expression of cholesterol bio-synthesis genes. (c). Fatty acid oxidation (FAO) can be upregulated by cMyc, depending on the cancer type as a means to counteract oxidative stress. ACC1/2: acetyl-CoA carboxylase 1/2, ACLY: ATP citrate lyase, ACS: acyl-CoA synthetase, α-KG: alpha-ketoglutarate, CoA: coenzyme A, CPT1: carnitine palmitoyltransferase 1, FADS: FA desaturases, FASN: fatty acid synthase, FPP: farne-syl-pyrophosphate, GLUT1: glucose transporter 1, HMG-CoA: hydroxy-methylglutaryl-CoA, HMGCS: hydroxy-methylglutaryl-CoA synthase, HMGCR: hydroxy-methylglutaryl-CoA reduc-tase, LD: lipid droplets, MUFA: monounsaturated fatty acids, PUFA: polyunsaturated fatty acids, SCD1: stearoyl-CoA desaturase 1, SOAT: sterol O-acyltransferase. The figure is created with Bio-Render.com (accessed on 26 March 2021). This figure is modified from Figure 1 in [78].