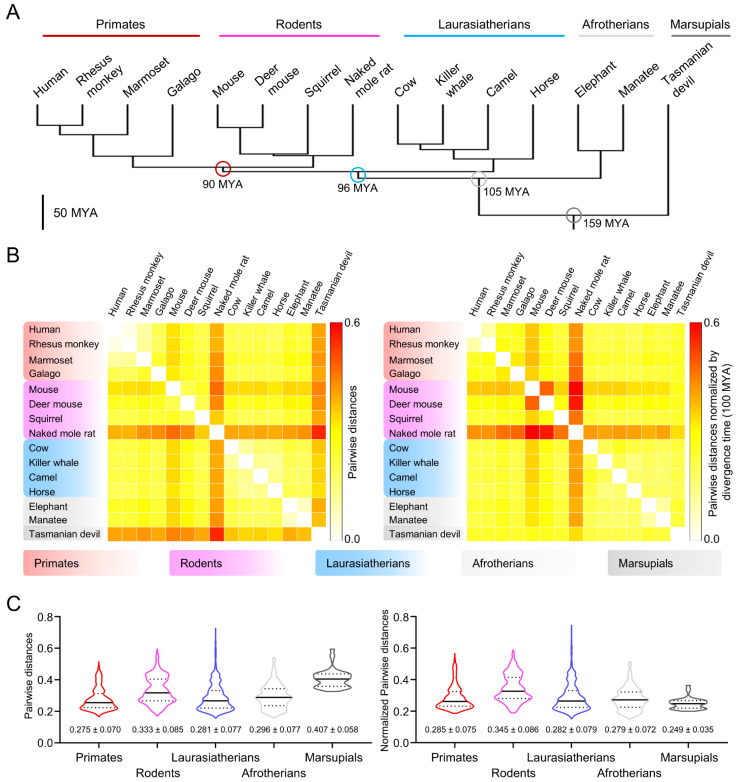
Figure 1.
Amino acid sequence comparison of mammalian CatSper orthologs shows that CatSper channel components in marsupials evolved slowly. (A) A phylogenetic tree of placental mammals. Divergence times of the clades, primates (human, rhesus monkey, marmoset, and galago), rodents (mouse, deer mouse, squirrel, and naked mole rat), Laurasiatherians (cow, killer whale, camel, and horse), Afrotherians (elephant and manatee), and a marsupial (Tasmanian devil) are marked with circles. Million years ago, MYA. (B) Pairwise distance analysis of CatSper subunits between placental mammals. Concatenated protein sequences of CatSper subunits (CatSper1-2-3-4-β-γ-δ-ε-ζ-EFCAB9) were used to calculate the pairwise distances between species. Raw (left) and normalized (right) pairwise distances are represented with heatmaps. Values of the raw pairwise distances were normalized by divergence times between two species (100 MYA = 1). Pairwise distances over 0.6 were set to 0.6 (red). (C) Interclade comparison of the CatSper orthologs. Distributions of the raw (left) and normalized (right) pairwise distances between species in different clades were represented in violin plots. Solid and dotted lines in each plot indicate median and quartile values, respectively. Mean ± SD values for the raw and normalized pairwise distances were marked at the bottom of each column. See also Figure S1.