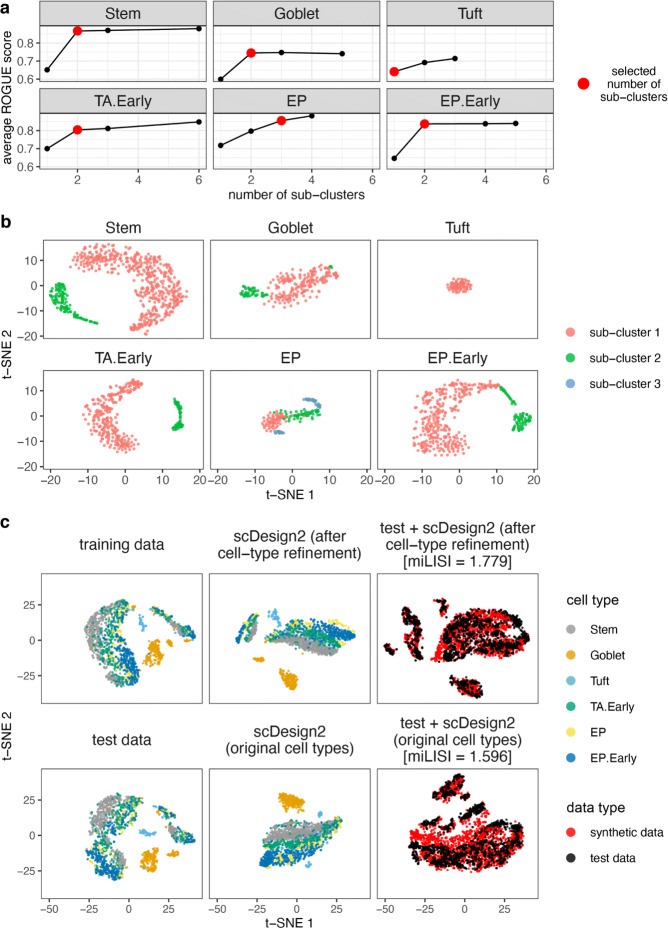
Fig. 5.
Application of ROGUE scores combined with dimensionality reduction plots to refine cell types before training scDesign2. This refinement approach is demonstrated on the 10x Genomics dataset. a In each cell type, the relationship between the average ROGUE score across sub-clusters and the number of sub-clusters. Before a ROGUE score is calculated for each sub-cluster, the Louvain clustering algorithm is applied to each cell type with a varying resolution parameter so that a varying number of sub-clusters is obtained. Based on how the average ROGUE score saturates, a number of sub-clusters is selected and marked in red for each cell type. b The t-SNE plots of each cell type with the sub-clusters, whose number is marked in a, labeled with distinct colors. c The t-SNE plots of training data (top left), test data (bottom left), synthetic data of scDesign2 trained with the refined sub-clusters (middle left) or the original cell types (middle bottom), and combination of test data with each set of synthetic data. Gene expression counts are transformed as log(1+count) before dimensionality reduction. We find that, after the cell type refinement, the simulated data of scDesign2 resemble the real data better, as indicated by the higher miLISI value