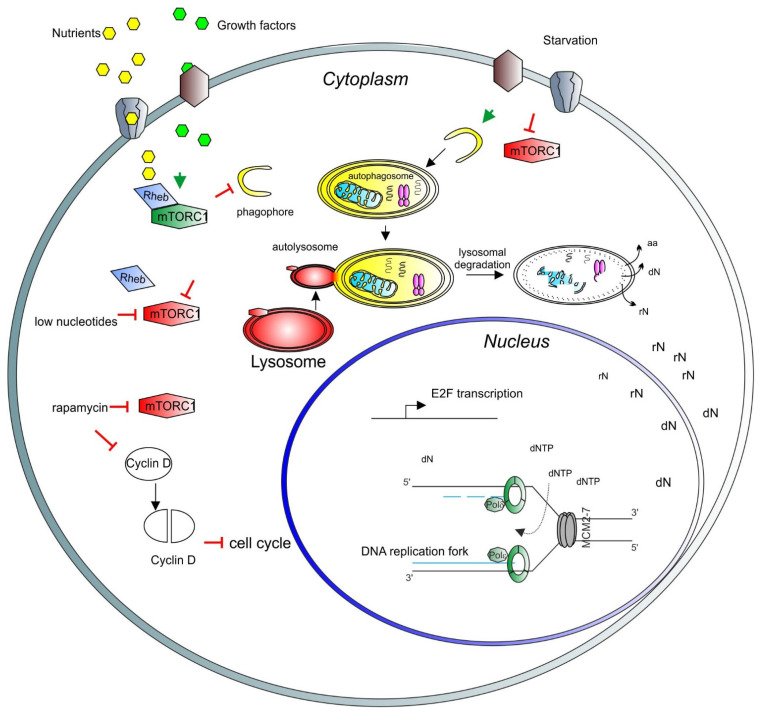
Figure 1.
A model depicting the role of mTORC1 in the cell cycle and nuclear activity regulation. Growth factors (light green hexagons) and nutrients (yellow hexagons) enter the cell through the membrane channels or bind and activate membrane receptors (brown and gray shapes), activating the mTORC1 complex (green long hexagon) via Rheb (blue rhomboid). Activated mTORC1 inhibits catabolic pathways, such as autophagy and lysosomal degradation. Upon starvation, mTORC1 is inhibited (red long hexagon) and autophagy can be triggered. Early stages of autophagy involve phagophore formation (yellow half-circle), followed by the formation of the autophagosome containing trapped cargo (yellow oval). The lysosome (red oval) then fuses with the autophagosome to form the autolysosome (red and yellow oval). Finally, the cargo is degraded, and recycled products are released to the cytosol for further use (aa = amino acids, rN = ribonucleosides, dN = deoxyribonucleosides). In the nucleus, nucleosides are converted to ribonucleotide triphosphates and deoxyribonucleotide triphosphates (dNTP) to be used for transcription and DNA synthesis, respectively. In the nucleus, a basic unit of DNA synthesis, called replication fork, is depicted. Newly synthesized DNA is shown as blue lines, the DNA polymerase complex is shown as green octagons and the DNA helicase complex MCM2-7 is shown as a grey barrel. A low level of nucleotides inhibits mTORC1. Rapamycin inhibits also mTORC1 activity, which induces degradation and accumulation of the D-type cyclins, negatively affecting the cell cycle progression. As a result, transcription of E2F and its target genes is inhibited. E2F activity is necessary for the transcription of genes involved in deoxynucleotide metabolism.