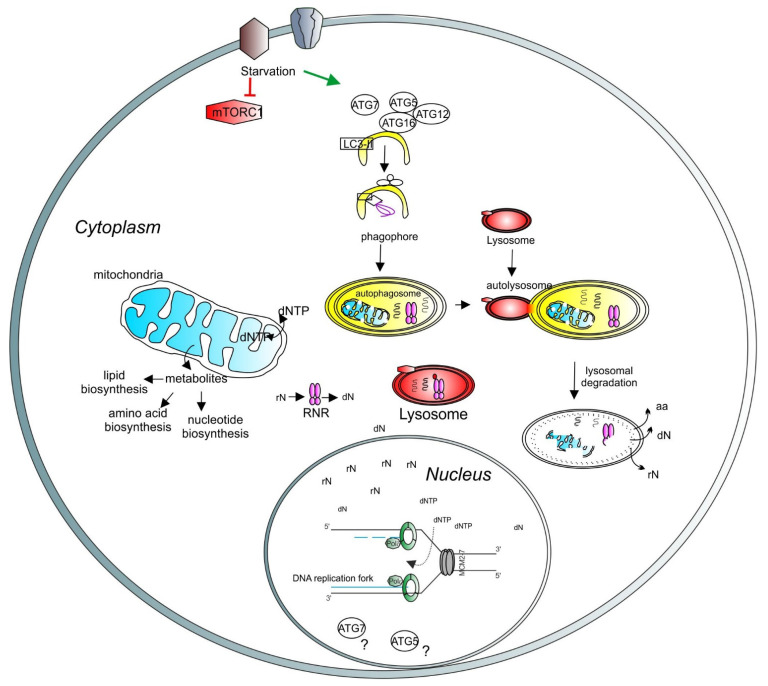
Figure 4.
A model depicting the role of lysosomes and autophagy in DNA synthesis and transcription. Lysosomal function is regulated by mTORC1 (red hexagons). Under starvation, autophagy is initiated by ATG proteins (empty circles) and LC3-related processes (empty square) to form the phagophore (yellow half-oval). The cargos are engulfed in the autophagosome, which then fuses with the lysosome to create the autolysosome, where the cargos are degraded. Recycled molecules are released to the cytoplasm for further use. Defective cellular organelles, such as mitochondria (blue shape), can be also targeted for degradation by autophagy. Healthy mitochondria contribute to the correct balance of intracellular metabolites. Intracellular nucleotide pools (ribonucleotides (rNTP) and deoxyribonucleotides (dNTP)) are important for maintaining adequate levels of DNA synthesis and transcription. Lysosomes can directly target the ribonucleotide reductase (RNR, pink shape) for degradation, thus regulating the deoxynucleotide level. A basic unit of DNA synthesis, called replication fork, is shown with DNA polymerases (green shapes) and the helicase MCM2–7 (grey shapes). The role of autophagy-related proteins, such as ATG5 and ATG7, in the regulation of the nuclear function requires further investigation (question marks).