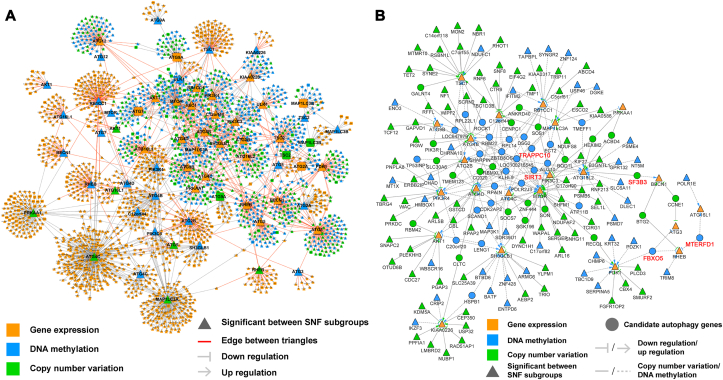
Figure 2.
Construction of the multi-omics network for BRCA using the lasso algorithm with TCGA data. (A) The direction of edges in the multi-omics network is consistently from MET/CNA to EXP. The arrows between nodes depict up- or down-regulation depending on the sign of the Pearson correlation coefficient of MET/CNA with EXP. Details are provided in Supporting Information Table S6. Nodes with significant differences in DNA MET/CNA frequencies and EXP between autophagic subgroups are denoted by triangles, and red edges between triangles indicate that the edges may be key interactions in the cancer-related core autophagy network. (B) Each node of the network is a statistically significant node. Details are provided in Supporting Information Table S7. Nodes linked with multiple core autophagic genes may have key roles in the regulation of the autophagy process and were identified as candidate autophagy targets. Five genes validated experimentally are highlighted in red.