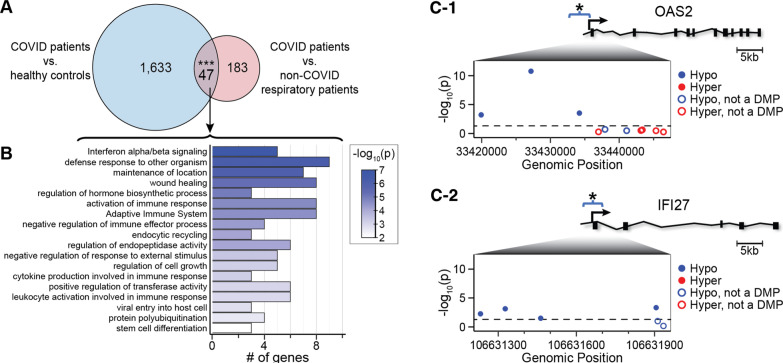
Fig. 4.
Overlap of COVID-19 DMR-associated genes in blood. A Venn diagram of the overlap of COVID-19 DMR-associated genes identified by comparison of DMRs between COVID-19 patients and healthy pre-pandemic controls, and DMRs between COVID-19 and non-COVID-19 respiratory illness patients on admission. Asterisks indicate overlap that is significant at p value < 0.001. Twenty-five of the 47 overlapping genes with DMRs encode proteins that participate in leukocyte viral defense, inflammation and immune responses. B Ontology analysis of the 47 overlapping genes with DMRs indicate a role in viral defense mechanisms. C Relative positions of COVID-19-associated DMRs in the promoter region of OAS2 (C-1) and IFI27 (C-2) with a schematic depicted for each gene. The relative positions of probes measuring methylation levels of CpG sites annotated to each gene with their genomic 5′-3′ positions are provided (inset panel; x-axis) versus the -log10 of the p value (y-axis). The p value < 0.05 is displayed as a black dashed line. Probes residing in a COVID-19-associated DMR are shown as hypo-methylation (blue dots) and hyper-methylation (red dots). Probes not meeting a p value < 0.05 at the individual CpG level are shown as hollow dots. These results indicate that the DMRs comprise a cluster of differentially methylated positions within days of SARS-CoV-2 infection