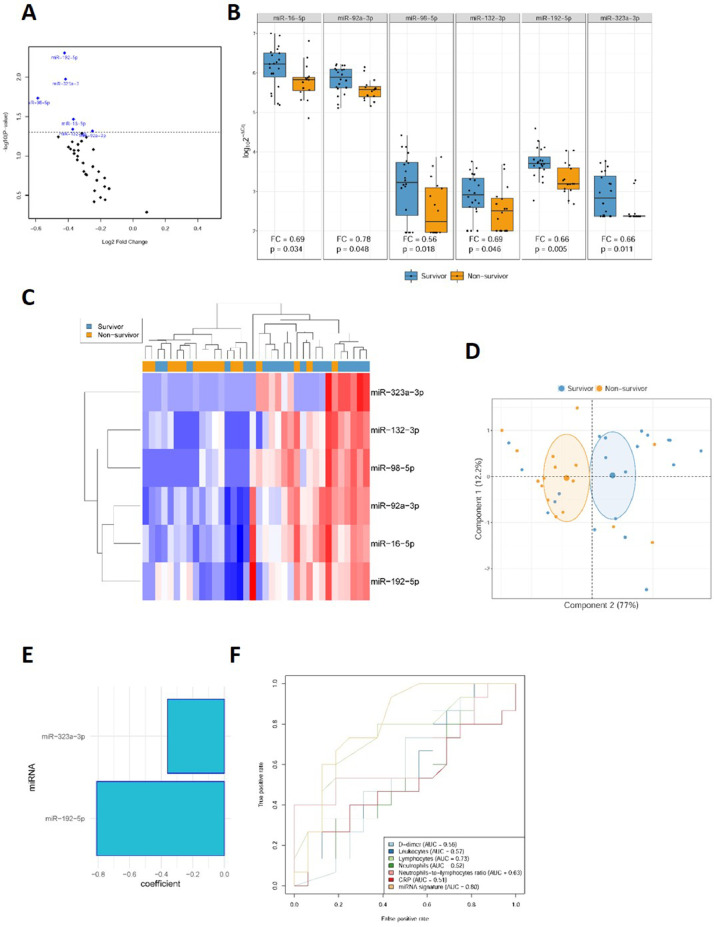
Fig 3.
Circulating microRNAs as biomarkers for ICU mortality in COVID-19 patients. A, Volcano plot of fold change and corresponding P-values for each microRNA after comparison of nonsurvivors and survivors (unadjusted). Each point represents one microRNA. Blue dots represent the microRNA candidates that showed significant differences; D, Box plot including plasma levels of microRNA candidates that showed differences between nonsurvivor and survivor patients. Between-group differences were analyzed using linear models for arrays. P-values describe the significance level for each comparison; C, Heat map showing the unsupervised hierarchical clustering. Each column represents a patient (nonsurvivor or survivor). Each row represents a microRNA. The color scale illustrates the relative expression level of microRNAs. The expression intensity of each microRNA in each sample varies from red to blue, which indicates relatively high or low expression, respectively. D, Principal component analysis. Each point represents a patient. E, Predictive model constructed using a variable selection process based on LASSO regression. miRNA levels were standardized prior to fitting the LASSO regression model. Estimated regression coefficients are shown. F, ROC curves for laboratory parameters and the microRNA signature. Expression levels were quantified by RT-qPCR. Relative quantification was performed using cel-miR-39-3p as the external standard. Relative quantification was performed using the 2−ΔCq method (ΔCq = CqmicroRNA-Cqcel-miR-39-3p). Expression levels were log-transformed for statistical purposes. microRNA levels are expressed as arbitrary units.