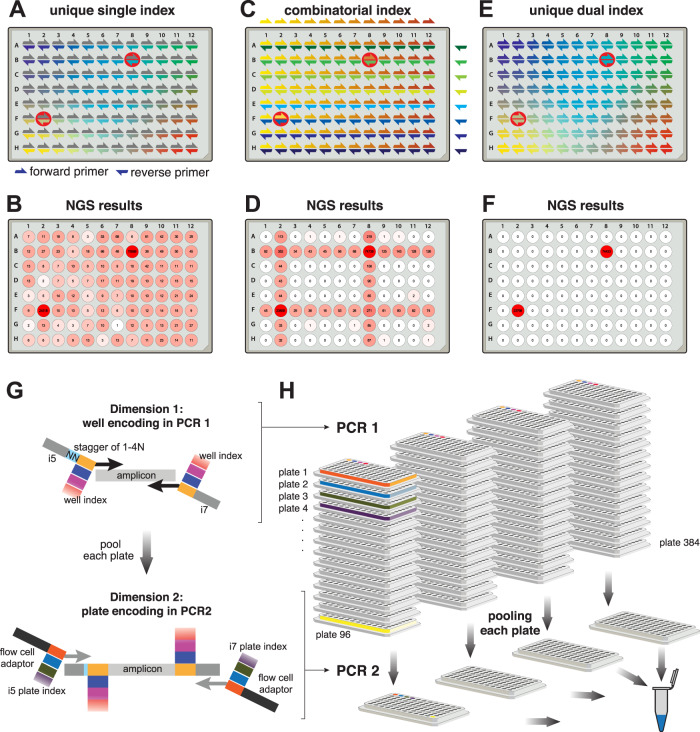
Fig. 2. Two-dimensional redundant dual indexing allows scaling to population-level testing.
A Scheme depicting 96-well-specific indices. These can be incorporated by forward or reverse primers. For the latter they can be incorporated during RT or PCR. Red circles highlight the positions into which synthetic SARS-CoV-2 RNA was added to test specificity of the indexing strategy. B N3 amplicon reads obtained by NGS and mapped to each well based on indices incorporated by reverse primers during PCR. Forward primer indices were present but disregarded in this analysis. Note the frequent misassignment to incorrect positions. C Scheme depicting combinatorial indexing. Each well is identified as a unique combination of a forward and a reverse index. D N3 amplicon reads mapped to wells based on combinatorial indexing as in C. To simulate combinatorial indexing the identical dataset as in B and F was used but for analysis, primers were treated in pools pointing to columns or rows. E Unique dual indexing is a redundant indexing method that encodes each well both by a unique forward and a unique reverse index. Thus, illegitimate recombination products between an amplicon and its associated indices can be bioinformatically rejected. F NGS result for the same dataset as in B and D analyzed with unique dual indices successfully filtered away all misassigned reads. G Two rounds of unique dual indexing (in two subsequent PCR reactions) can be used to index first wells and then plates, effectively achieving combinatorial (multiplicative) scaling. Colored boxes represent indices. H Illustration of the PCR workflow. RT and PCR1 are performed on all samples individually, adding well-specific indices. Subsequently, each plate is pooled to one well of a new plate, reactions are treated with Exostar to remove excess primers and a second PCR is done, adding plate-specific indices and the Illumina flow cell adaptors. Our currently used and validated index set allows pooling of up to 36,864 samples (96 in PCR1 × 384 in PCR2). Finally, PCR2 amplicons are pooled, gel purified, and sequenced on any Illumina platform.