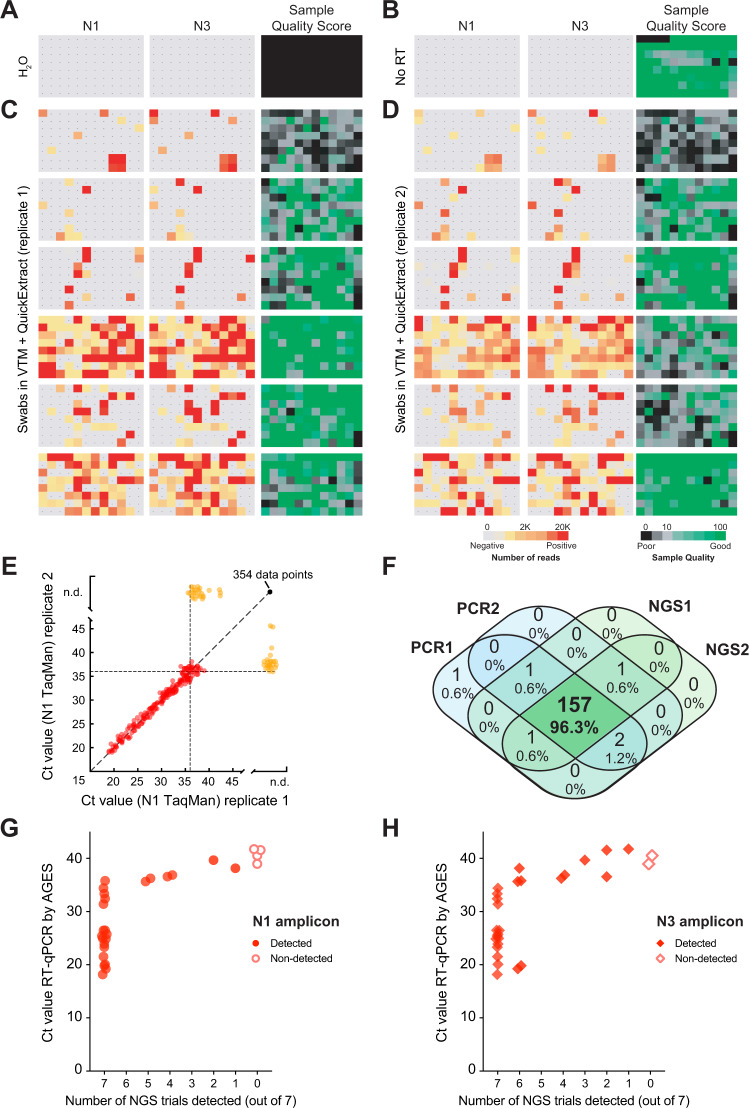
Fig. 4. SARSeq robustly detects SARS-CoV-2 in patient samples.
A H2O control plate of SARSeq with three primer pairs, namely N1, N3, and ribosome in the presence of RTC as well as N1 and N3 scavenger spike-ins. B SARSeq on a gargle-QuickExtract plate in the absence of reverse transcriptase. No false-positive wells were detected. Sample QC scores high due to the absence of RTC reads while DNA templated ribosomal amplicon is observed. C Analysis of 576 samples obtained by swab and collected in VTM. D Independent replicate of C based on the same inactivated patient swab. Color codes depict read counts and sample quality score for C and D. E Two independent N1 TaqMan qPCR runs on samples used for C and D. Ct values of both runs are plotted against each other. Color code: red: sample scoring positive in both qPCR replicates, orange: sample scoring once, black: sample scoring negative. Stochastic and latest detection of SARS-CoV-2 at cycle 36. F Venn diagram of reproducibility for all samples scoring positive with Ct 36 or less for at least one qPCR replicate. G, H Comparison of NGS results by SARSeq to Ct values obtained by diagnostic qPCR. A set of 90 samples (including swabs and gargle in different buffers) was used for RNA extraction and qPCR measurements and in parallel aliquots of these samples were mixed either with QuickExtract or TCEP/EDTA and measured by SARSeq in triplicates and quadruplicates, respectively. We report the number of replicates in which we called a sample positive by NGS (with the N1 or N3 amplicon) relative to the qPCR Ct values. Not shown are 63 samples that were negative by qPCR and NGS.