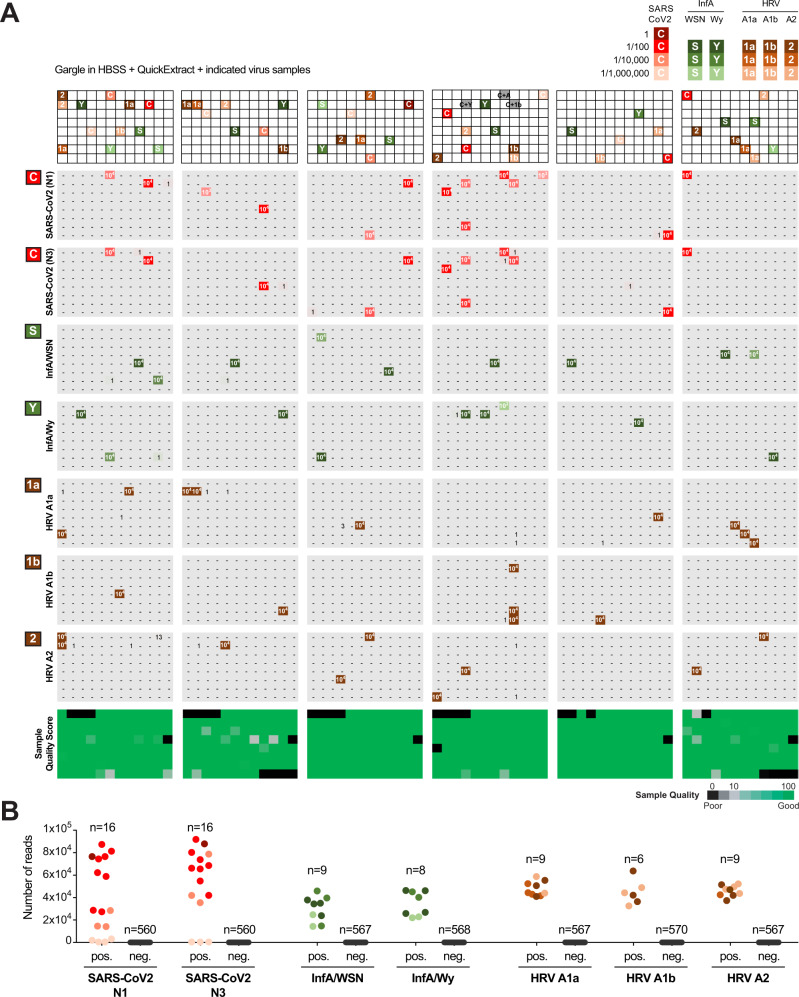
Fig. 5. SARSeq can detect multiple respiratory viruses in a single reaction.
A Six 96-well plates filled with gargle in HBSS and inactivated in QuickExtract from our in-house pipeline were spiked with RNA from various respiratory viruses. For SARS-CoV-2, a positive gargle sample with Ct value of 30 was diluted as indicated. RNA for all other viruses was obtained from HEK cells 48 h after infection with the virus. Dilution indicated by voluminometric ratio. SARSeq was performed with six primer pairs in one reaction, namely N1 and N3 for SARS-CoV-2, Influenza A virus, human rhinovirus (HRV), and the ribosome. Influenza A substrains and HRV substrains were distinguished based on amplicon sequence variants. B Analysis of false-positive and false-negative rate of the experiment in panel A. As expected, 1:100,000 dilutions of the SARS-CoV-2 sample with a Ct of 30 were missed. All other positive samples were detected for all viral spike-ins.