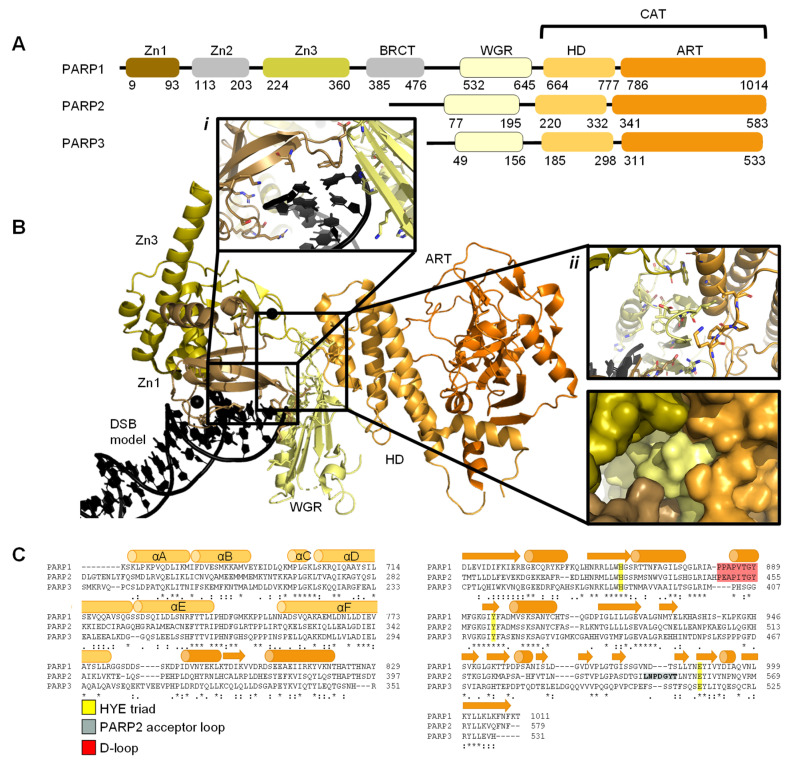
Figure 1.
Overview of PARP1, PARP2, and PARP3. (A). Schematic overview of the domains of PARP1, PARP2, and PARP3, with amino acid numbers indicating domain boundaries corresponding to Uniprot entries P09874, Q9UGN5, and Q9Y6F1; (B). Crystal structure of PARP1 bound to a double-strand DNA molecule representing one end of a double-strand break (DSB) (Protein Data Bank (PDB) entry 4DQY [34]). Domain colours correspond to A. Inset i: interactions of Zn1 and WGR with a DSB model. Inset ii: interdomain interactions between Zn1, Zn3, WGR, and HD shown as cartoons (top) and surface representations (bottom). All figures in this review containing protein structures were generated in PyMOL [43]; (C). Structure-based sequence alignment of CAT domains of PARP1, PARP2, and PARP3 was generated in PyMOL [43]. Sequence alignment was generated in Clustal Omega [44] provided by EMBL-EBI [45]. Secondary structure is represented by cylinders as α-helices and arrows as β-sheets, respectively. Sequence conservation is shown, where an asterisk represents fully conserved, a colon represents strongly similar properties and a period represents weakly similar properties. The conserved histidine, tyrosine, glutamic acid (HYE) triad is shown in yellow, the PARP2 acceptor loop in grey, and the D-loop in red.