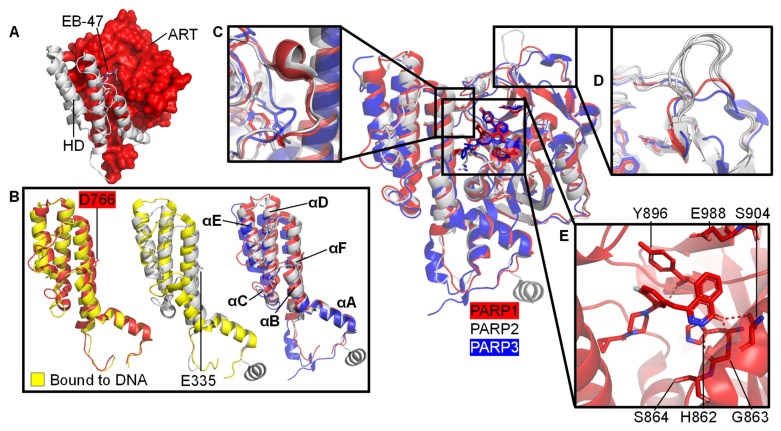
Figure 3.
Structural details of the CAT domains of PARP1-3. Central image shows a structural superposition of the ART domains of PARP1 (red, olaparib-bound, PDB 7AAD [109]), PARP2 (white, olaparib-bound, PDB 4TVJ [115]), and PARP3 (blue, ME0354-bound, PDB 4GV2 [116]) with highlighted features. (A) A folded HD (white) occludes the NAD+ binding pocket in the PARP1 ART domain (red, PDB 7AAB, PARP1 with NAD+ analogue EB-47 bound [109]); (B) Subtle movement of parts of the HD domains in response to DNA binding after superposition of the ART domain only. Left: HD of apo PARP1 before (red, PDB 7AAA [109]) and after (yellow, PDB 4DQY [34]) DSB model binding. Middle: HD of PARP2 before (white, PDB 4TVJ [115]) and after (yellow, PDB 6X0N [53]) DSB model binding. Right: HDs of PARP1, PARP2, and PARP3 in complex with inhibitors as specified above with annotation of the helices; no DNA binding; (C) Close-up of the D-loop of PARP1 and PARP2, which is absent in PARP3; (D) The acceptor loop of five representative PARP2 structures (white: PDBs 4ZZX [117], 6X0L, 6X0M, 6X0N [53], 4TVJ [115]) contains a unique insertion compared to PARP1 (red, PDB 7AAD, bound to olaparib [109]) or PARP3 (blue, PDB 4GV2, bound to ME0354 [116]). This is suggested to increase branching of PAR chains by PARP2 [112]; (E) Close-up of the NAD+ binding pocket of PARP1 (red, olaparib-bound, PDB 7AAD [109]). The catalytic triad involving H862, Y896, and E988 and the residues interacting with PARPi, G863-S864, and S904, are indicated.