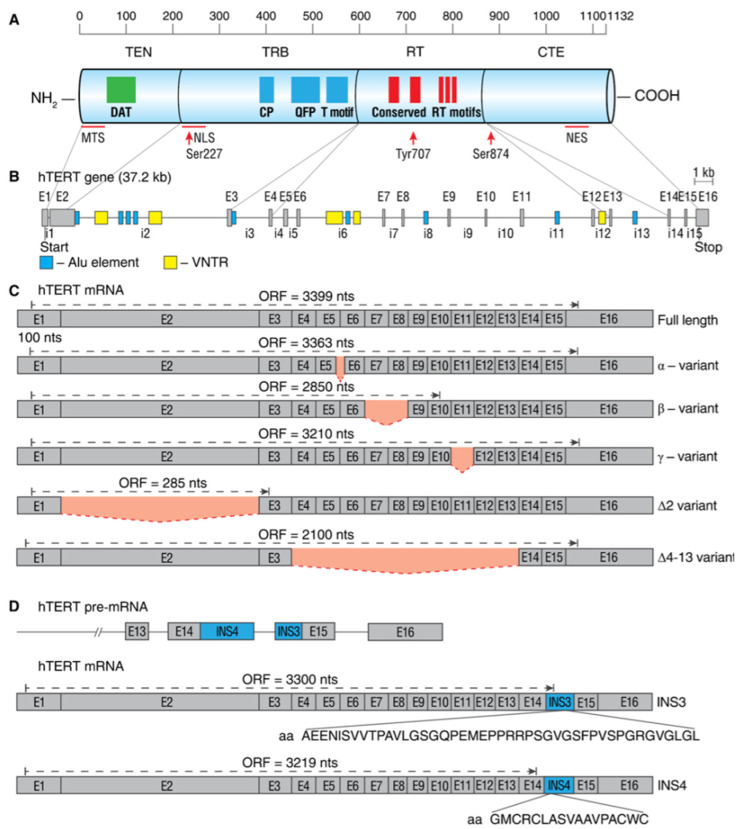
Figure 2.
Schematic map of hTERT protein, gene, and commonly studied mRNA splice variants. (A) Linear structure of 1132-amino acid hTERT protein and known domains and motifs are shown. The following active elements are responsible for intracellular relocalization of hTERT: MTS, mitochondrial targeting sequences; NLS, nuclear localization signal; Ser227, Serine 227 for phosphorylation by Akt; Tyr770, Tyrosine 770 for phosphorylation by Src1; Ser824, Serine 824 for phosphorylation by Akt; NES, nuclear export signal for binding with CRM1. (B) Structure of hTERT gene exons (E1–E16) and introns (i1–i15). Positions of Alu elements and variable number tandem repeats (VNTRs) are shown as dark blue and yellow boxes, respectively. Lines link exons and the domains they encode. (C) Common alternatively spliced variants with deletions are shown below the wild-type, the full-length mRNA. Predicted open reading frame (ORF) for each mRNA is indicated. (D) Common alternatively spliced variants that include insertions INS3 and INS4 and the amino acids that are encoded by them.