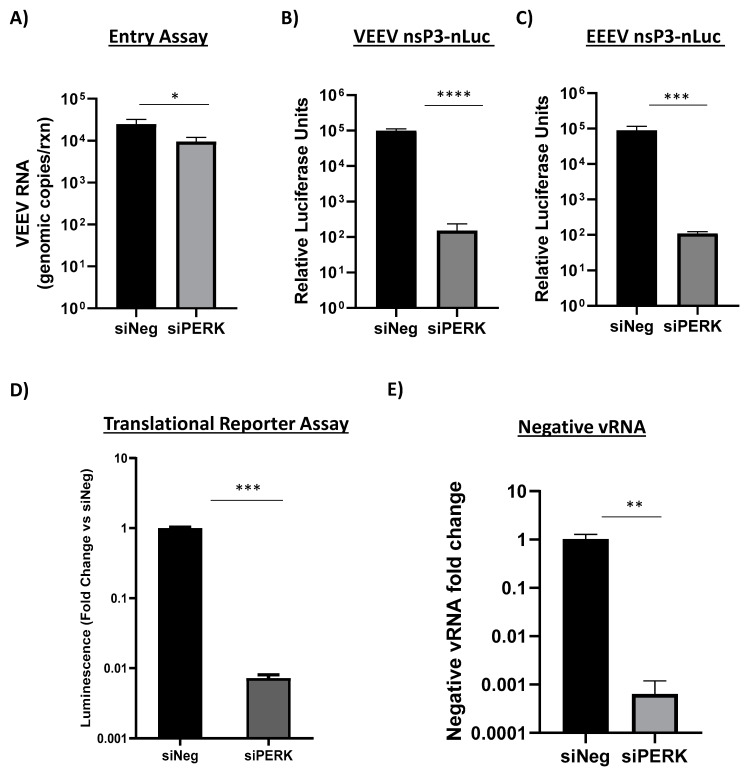
Figure 6.
Loss of PERK signaling minimally impacts VEEV entry but inhibits the translation of incoming alphavirus genomes and production of negative-stranded viral RNA. (A) Primary astrocytes were transfected with 100 nM of siNeg or siPERK siRNAs. At 48 h post transfection, cells were infected with VEEV (MOI 5) and RNA collected 1 hpi. Viral genomic copies were determined by RT-qPCR. Results are displayed as genomic copies in logarithmic scale. Data are expressed as the mean ± SD (n = 3). * p ≤ 0.05 using Student’s t-test. (B,C) Primary astrocytes were transfected with 100 nM of siNeg or siPERK siRNAs. At 48 h post transfection, cells were infected with VEEV nsP3-nLuc (panel B) or EEEV nsP3-nLuc at MOI of 5 (panel C). At 18 hpi, luminescence was measured using Promega’s Nano-Glo Luciferase Assay system. Data are expressed as the mean ± SD (n = 5). *** p ≤ 0.001, **** p ≤ 0.0001. (D) Transfected cells were electroporated with translation reporter RNAs. Cells were lysed 2 h post electroporation and luciferase activity was measured. siPERK transfected and VEEV reporter RNA electroporated cells are expressed as RLUs (relative luminescence units) per µg protein expressed as a fold change over siNeg transfected and VEEV reporter RNA electroporated cells. Data are expressed as the mean ± SD (n = 3). *** p ≤ 0.001. (E) Primary astrocytes were transfected with 100 nM of siNeg or siPERK siRNAs. At 48 h post transfection, cells were infected with VEEV TrD at MOI of 5. At 18 hpi, RNA was extracted and negative-stranded viral RNA detected via RT-qPCR. siNeg samples were set to a fold change of 1. Data are expressed as the mean ± SD (n = 3). ** p ≤ 0.01.