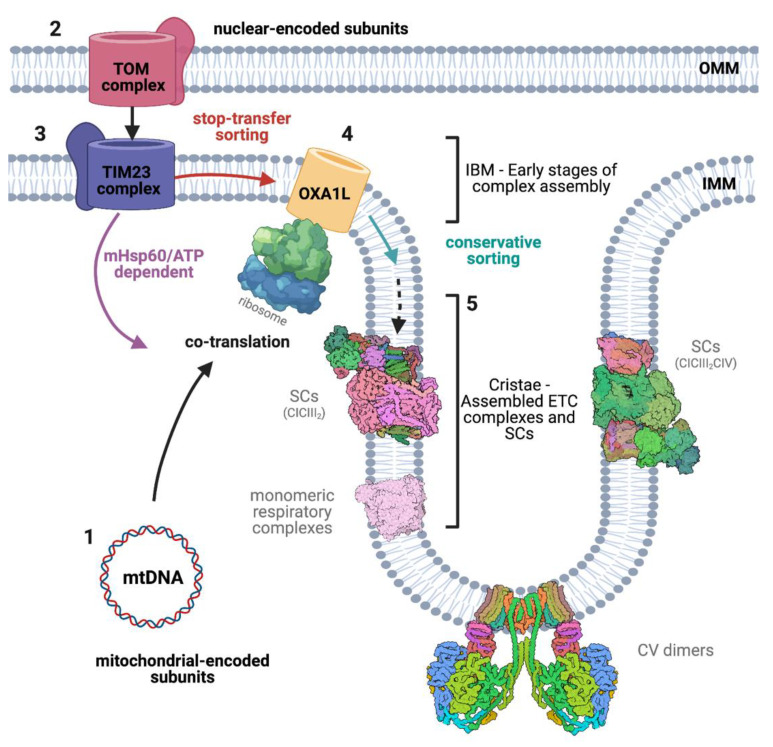
Figure 2.
Spatial orchestration of mitochondrial respiratory complexes import and assembly and their organisation in the IMM. ETC complexes I, III, IV, and V are composed of both mitochondrial and nuclear-encoded subunits. Transcripts from the mitochondrial genome (1) are co-translated by mitochondrial ribosomes (here depicted as a simple arrow for clarity) and proteins inserted in the IMM via OXA1L. These newly synthesised proteins are then assembled together with the nuclear-encoded subunits, which are imported primarily through the TOM/TIM23 (2,3) complex. Additionally, proteins carrying a hydrophobic segment downstream of the MTS are arrested in the Tim23 channel and laterally inserted into the IMM through a stop-transfer sorting mechanism acquiring a Nin/Cout topology. Proteins with a Nout/Cin topology are instead fully imported and inserted into the IMM from the matrix side through a process known as conservative sorting, involving OXA1L (4). The import and insertion of these subunits in the IMM take place predominantly in IBM, a section of the IMM that runs parallel to the OMM. Then, the ETC subunits undergo a series of post-translational modifications and are incorporated in a nascent enzyme, often due to the interaction with assembly factors or chaperons. This process can occur in the monomeric enzymes and/or in the high-order SCs. Fully assembled enzymes and SCs are enriched in the cristae region of the IMM (5). Note: the size of monomeric respiratory complexes, supercomplexes, import machineries, and ribosomes are not to scale.