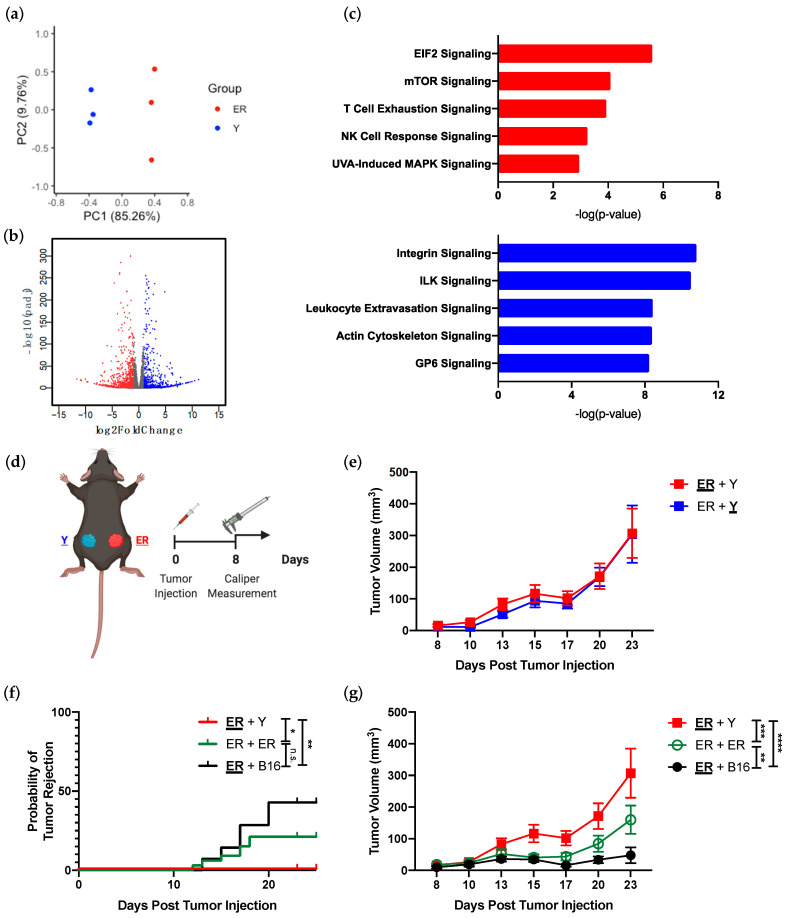
Figure 1.
YUMMER (ER) and YUMM (Y) cell lines have different transcriptomes in vitro and establish synchronous melanoma tumors in vivo. (a) PCA plot separating overall transcriptomic differences between ER and Y cell lines in vitro. Notice how the two cell lines generate distinct groupings along the PC1 axis. (b) Volcano plot highlighting differentially expressed genes in ER (red) vs. Y (blue) cell lines. Colored genes are statistically significant with adjusted p-value < 0.05 and log 2-fold change > |1|. (c) Top-most upregulated pathways in ER and Y cell lines as identified by IPA analysis. Red bars indicate pathways upregulated in ER and blue bars indicate pathways upregulated in Y cells. (d) Synchronous murine melanoma model schematic. The analyzed tumor in the synchronous model is underlined and bolded in subsequent figures. A table of tumor combination can be found in Supplementary Table S1. (e) Growth curves of individual ER (ER + Y) and Y (ER + Y) tumors in synchronous melanoma mice. (f) Percent of ER tumors rejected and growth curve (g) of synchronous ER tumors from ER + Y, ER + ER, and ER + B16 mice. Data (mean ± SEM) in (e–g) are pooled, from 3–5 mice/group/experiment, and representative of at least 2 independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, n.s. not significant.