Extended Data Figure 2.
Characterization of gene expression in old Paneth and ISCs
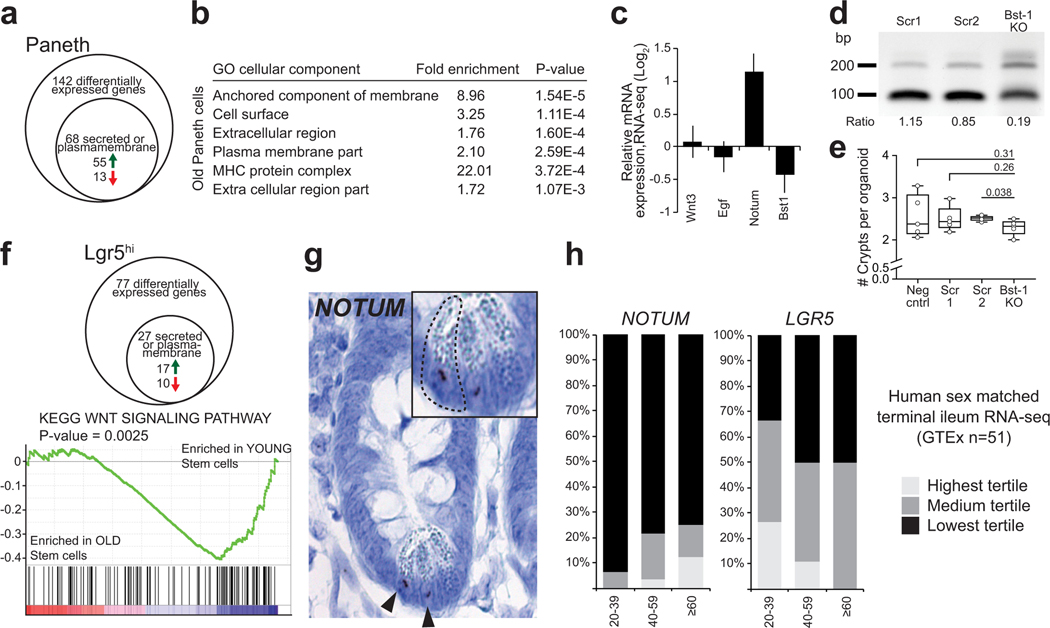
a, Venn-diagram of gene expression changes in old Paneth cells. (n=5 animals in old, n=4 animals in young) b, List of Gene Ontology (GO) terms with the highest enrichment among genes deregulated in old Paneth cells, Fisher’s exact test, no correction for multiple testing. c, Expression of stem cell maintaining factors Wnt3, Egf, and of Notum and Bst-1 in old Paneth cells (RNA-seq). Values show fold change in comparison to young Paneth cells. (n=5 animals in old, n=4 animals in young). d, Gene editing of Bst-1 confirmed by PCR strategy with primers flanking the editing site (191bp product) and hitting the edited site (89bp product). Representative agarose gel image is shown. Experiment repeated once to validate the organoid line used in Extended Data Fig. 2e. e, Regenerative growth of Bst-1 KO intestinal organoids. Organoids were quantified 2 days after subculturing (n=5 repeated experiments with the same organoid line). f, Venn-diagram of gene expression changes in old Lgr5hi stem cells. GSEA preranked analysis of old versus young Lgr5hi stem cells for the gene list “KEGG WNT SIGNALING PATHWAY”. Nominal P-value is shown. (n=3 animals per age group). g, RNA-scope for NOTUM mRNA (brown) in human jejunal section. Expression seen exclusively in Paneth cells (arrowheads and inset). Experiment repeated twice with similar results in independent samples. h, Expression of human NOTUM and LGR5 from terminal ileal samples of GTEx Consortium (n=51 sex matched samples). Expression range is divided to three equal-sized tertiles. Unless otherwise indicated, line at Box-and-Whisker -plots represents median, box interquartile range and whiskers range. All other data are represented as mean +/− s.d. and conditions compared with two-tailed unpaired Student’s t-test, exact P-values shown in corresponding panels. P-values < 0.05 were considered significant. For gel source data see Supplementary Fig. 3.