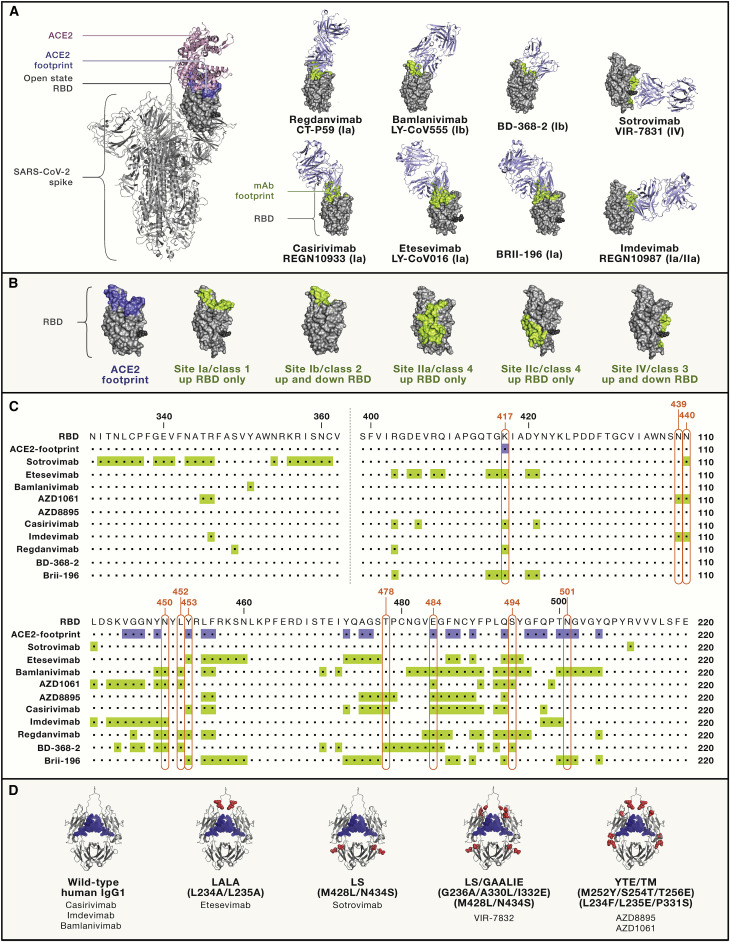
Figure 2.
Fab-RBD complexes, epitopes, and Fc mutations of clinically relevant mAbs
(A) Full spike (S) of SARS-CoV-2 (PDB: 6ZGG) is shown on the left, where ACE2 (pink cartoon) is modeled on the open-state RBD (gray space-filling model) (ACE-2 PDB: 6M0J); the structures of eight Fab-RBD complexes were determined by a combination of X-ray crystallography and/or cryoelectron microscopy (cryo-EM) analysis (Fabs shown as light-blue cartoons and RBD orientation fixed in the upward state shown on S trimer [left image]; see Table 1 for PDB accession numbers). ACE2 and mAb footprints are shown in blue and light green, respectively. Footprints on RBDs were defined according to 5 Å distance from ACE2- or mAb-contacting residues. The stem of N343 glycan is shown as a black sphere. mAbs are labeled using both their original and generic (nonproprietary) names.
(B) Antigenic sites nomenclature (Ia–IV versus classes 1–4) according to Barnes et al., 2020a, Cohen et al. (2021), and Piccoli et al. (2020) and colored as in (A).
(C) Sequences of the full RBD of SARS-CoV-2 (Wuhan-1 strain), where ACE2 and mAb footprints are highlighted in blue and light green, respectively, as in (A) and (B). The key RBD mutations found in VOCs/VOIs are boxed in red.
(D) Structural representation of human IgG1 Fc where amino acids changed in COVID-19 mAbs in late development are shown as red spheres. N297-bound glycans are shown as blue spheres. List of Fc abbreviations: LALA, L234A/L235A (Hezareh et al., 2001; Xu et al., 2000); GAALIE, G236A/A330L/I332E (Weitzenfeld et al., 2019); YTE, M252Y/S254T/T256E (Dall’Acqua et al., 2002); LS: M428L/N434S (Zalevsky et al., 2010); TM, triple mutant in Fc, L234F/L235E/P331S (Oganesyan et al., 2008).