Figure 2.
Slow network-wide activation of GoCs
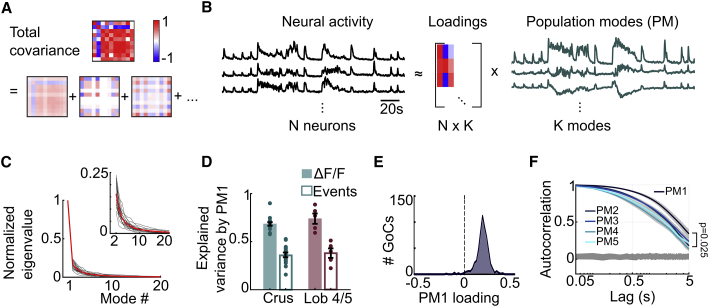
(A) Illustration of how correlations between GoCs were quantified by decomposing total covariance into contributions from different modes.
(B) Illustration of extraction of population modes from neural activity using principal-component analysis. Time-varying neural activity (left), the loading matrix (center), and activity along population modes (right). The loading matrix gives the weight of mode k for neuron n.
(C) Distribution of eigenvalues for the population covariance matrix (for ΔF/F), normalized by the maximum eigenvalue in each session. Black lines are individual sessions (n = 21 sessions, N = 9 mice), solid red line is the mean across sessions. Inset shows the expanded axis without population mode 1 (PM1).
(D) Cross-validated explained variance by first PM alone for ΔF/F (solid) and events (open) in Crus I/II (cyan) and Lob IV/V (magenta). Points indicate mean across neurons on an individual session; shaded bar and error bar indicate mean across sessions and SEM, respectively.
(E) Distribution of loadings for top mode (PM1, n = 582 neurons/N = 9).
(F) Autocorrelogram for the first 5 population modes (darkest PM1, lightest PM5); shaded areas denote SEM across sessions. Gray area at bottom shows the 95% confidence interval for the time-shuffled control. p value reported at 5-s lag (PM1 versus non-PM1); Wilcoxon signed rank test, n = 21/N = 9.
See also Figure S3.