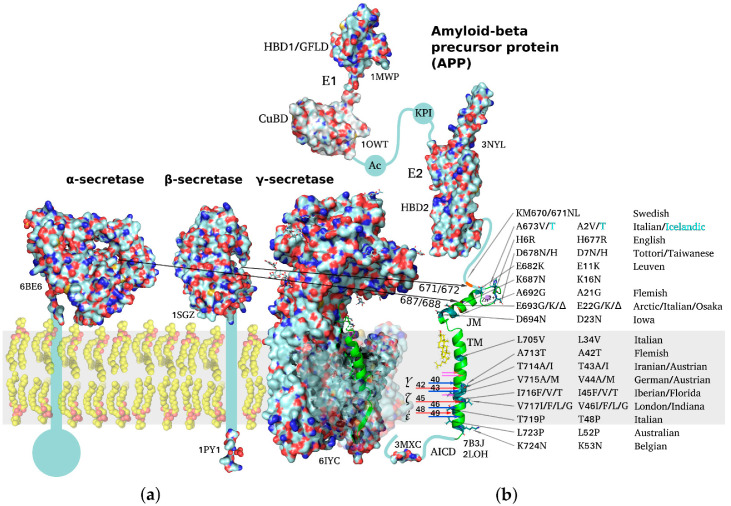
Figure 1.
Schematic representation of structures of amyloid precursor protein and α-, β-, and γ-secretases responsible for Aβ production. PDB accession number is indicated for each molecule throughout the figure. (a) Molecular surfaces of the structural elements of α-, β-, and γ-secretases. Single-span transmembrane (TM) domains of α- and β-secretases are shown as bars where high-resolution structure is unavailable. (b) Full-length APP structure based on individually resolved structures of its parts: flexible intracellular C-terminal domain (AICD), TM domain, connected through a flexible extracellular juxtamembrane (JM) region (containing Aβ metal-binding domain) to an ectodomain consisting of (i) E1 subunit including a cysteine-rich growth factor-like domain (HBD1/GFLD) and a copper/zinc-binding domain (CuBD), an acidic region (Ac), a Kunitz-type protease inhibitor domain (KPI), and (ii) E2 subunit with a second heparin-binding domain (HBD2). Resolved domain structures are shown as ribbon diagrams, and unstructured flexible connecting loops are shown as solid lines. The familial mutations attributed to increased risk or earlier age of AD development are shown in black on the TM and JM segments. A673T mutation decreasing APP proteolysis by β-secretases is highlighted in cyan. Sites of cleavage by α-, β-, and γ-secretases are indicated by arrows color-coded to distinguish between two alternative cleavage cascades generating Aβ1–42 and Aβ1–40 peptides (48 > 45 > 42 vs. 49 > 46 > 43 > 40). Cholesterol molecule interacting with the N-terminal part of APP TM helix is shown. The inset demonstrating the helical APP TM domain (in green) with a C-terminal turn (3 a.a. residues) unfolded into a β-strand is shown in the γ-secretase active center.