Figure 2.
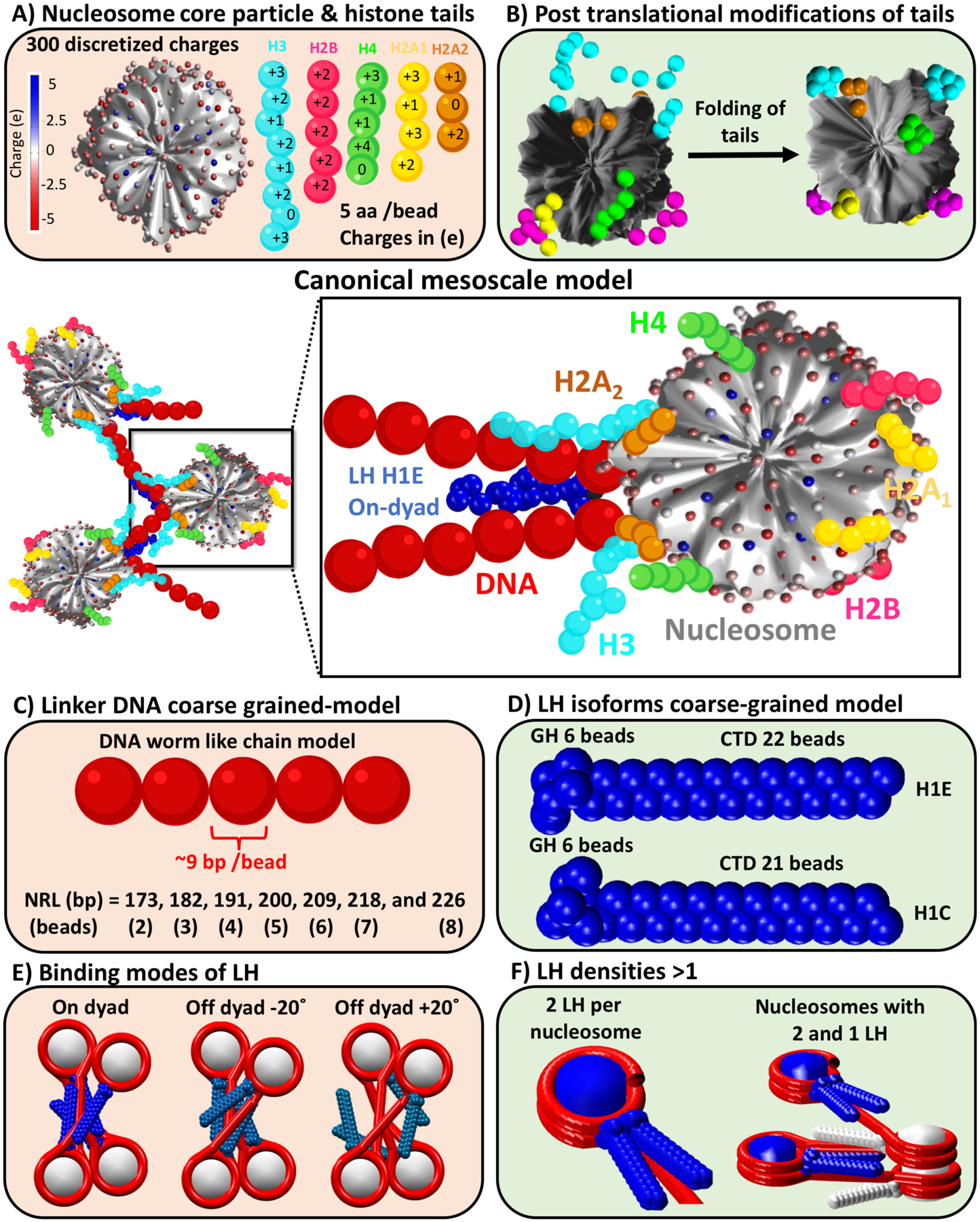
Our canonical chromatin mesoscale model at center with all its components detailed as follows: A) Rigid nucleosome core particle (NCP) modeled by 300 discrete charges determined with the DiSCO algorithm (90), along with flexible histone tails (H3 N-tail in cyan, H2B N-tail in magenta, H4 N-tail in green, H2A N-tail in yellow, and H2A C-tail in orange), coarse-grained as 5 amino acids per bead with charges also determined by DiSCO (90, 173); B) Nucleosome with wild type (left) and with folded histone tails (right) containing lysine acetylation modeled by increased stretching, bending, and torsional intertail-bead force constants by a factor of 100 (95); C) Linker DNA modeled as a worm-like chain polymer, coarse-grained as ~9 bp per bead. The nucleosome repeat lengths (NRLs) of 147 bp plus linker DNA length in bp are modeled by 2, 3, 4, 5, 6, 7, and 8 beads to mimic NRLs = 173, 182, 191, 200, 209, 218, and 226 bp or DNA linker lengths = 26, 35, 44, 53, 62, 71, and 79 bp, respectively (202); D) Linker histone (LH) isoforms H1E and H1C, modeled with 22 and 21 beads, respectively (5 amino acids per bead) for the CTDs and 6 beads for GHs with their charges determined by DiSCO (90, 93, 94); E) On and off-dyad (+20° and −20°) binding modes for the LHs (93); and F) Chromatosome with 2 LH bound (left) and tetranucleosome fiber with a density of 1.5 LH per nucleosome (right), where nucleosomes with 2 LH are colored blue and with 1 LH are colored white.
