Figure 3.
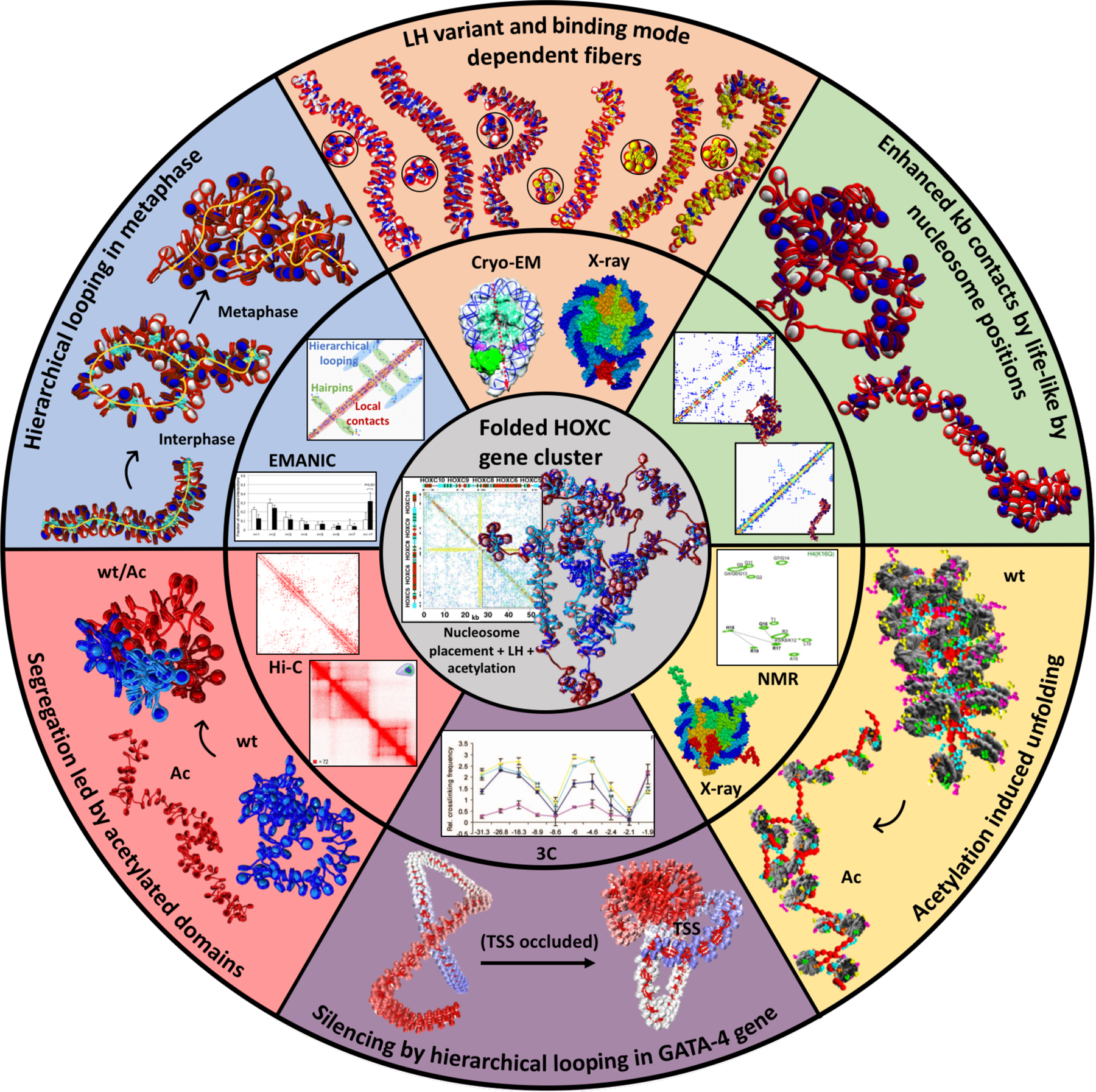
Mesoscale modeling studies emphasizing the interplay between experiment (middle ring) and modeling (outer ring), along with crucial internal/external fiber parameters that direct gene folding, as demonstrated for the HOXC gene cluster (center). Clockwise from blue slice: Hierarchical looping in metaphase chromosomes. Computed fiber structures for terminally differentiated cells (1 LH per nucleosome), interphase chromatin (0.5 LH per nucleosome), and metaphase chromatin (no LH), with LHs in turquoise explain nucleosome contacts determined by the EM-assisted nucleosome interaction capture (EMANIC) technique for metaphase (black bars) and interphase (white bars) chromatin in situ, by the hierarchical looping folding motif, also evident in the accompanying computed contact matrix (70). Peach slice: LH variant and binding mode dependent fibers. The chromatin fiber topologies (top and side views) are sensitive to different combinations of LH variant, binding mode, and density. The six fibers, from left to right, correspond to: ρ = 1, 100 H1E on-dyad; ρ = 1, 100 H1E −20°; ρ = 1, 100 H1E +20°; ρ = 1.3, 100 H1E +20° and 30 H1C on-dyad; ρ = 1.6, 100 H1E +20° and 60 H1C on-dyad; and ρ = 1.6, 40 H1E +20°, 60 H1E +20°, and 60 H1C −20°, where nucleosomes containing two LH bound are colored in yellow (93). The crystal structures of a chromatosome with LH (in red) bound on-dyad (PDBID: 4QLC (203)) and Cryo-EM chromatosome with LH bound off-dyad (in green) were used to generate our two binding modes. The cryo-EM image was adapted with permission from (35). Green slice: Enhanced kb contacts by life-like nucleosome positions. Fibers with life-like linker lengths and nucleosomes free regions (NFRs) generate many more long-range kb contacts compared to uniform linker-length fibers. This is evident by the folded fibers and the associated contact maps, which indicate hierarchical looping in life-like fibers compared to mostly short-range interactions in uniform fibers (99). Yellow slice: Acetylation induced unfolding. Compared to a wild type chromatin (wt), the fiber with acetylated histone tails (Ac) drives global unfolding due to lack of stabilizing internucleosome interactions (95). The experimental NMR spectra of the core histone H4 K16Q were used to validate the acetylated tail structures obtained by all-atom molecular dynamics simulations. Adapted with permission from (204). The crystal structure of the nucleosome (PDB: 1KX5 (30)) was used to construct dinucleosomes in our multiscale modeling (95). Violet slice: Gene silencing by hierarchical looping in the GATA-4 gene. The experimental 3C contacts in 4 different cells (blue, UT cells; violet, DT cells; yellow, HCT116 cells; and light blue, DKO cells) were used as constraints in our GATA-4 gene model, with image adapted from (181). The representative unfolded (left) and folded (right) GATA-4 gene structures suggest how folding by hierarchical looping would silence the transcription start site (TSS) of the gene (178). Pink slice: Segregation induced by acetylated domains. Intrinsic compartments of acetylated (Ac)/wild type (wt) segments form at the kb level for a mixed fiber construct (50% wt and 50% Ac) compared to fibers with 100% wt (blue) and 100% Ac (red). Both the Hi-C contact map of a segment of human Chr3 and our computed contact map of the alternating fiber construct show segregation patterns. The Hi-C contact map image was adapted with permission from (96). Gray inner portion: Folded HOXC gene cluster. All previous internal/external fiber parameters, including nucleosome positions, LH binding positions, and acetylation islands are combined to fold in silico the HOXC gene cluster and reveal a contact hub (101); see also commentary on our work in (198). The contact map of the folded HOXC gene cluster unravels a central interaction between two domains: LH-rich (top left of contact map) and acetylation-rich regions (bottom right).
