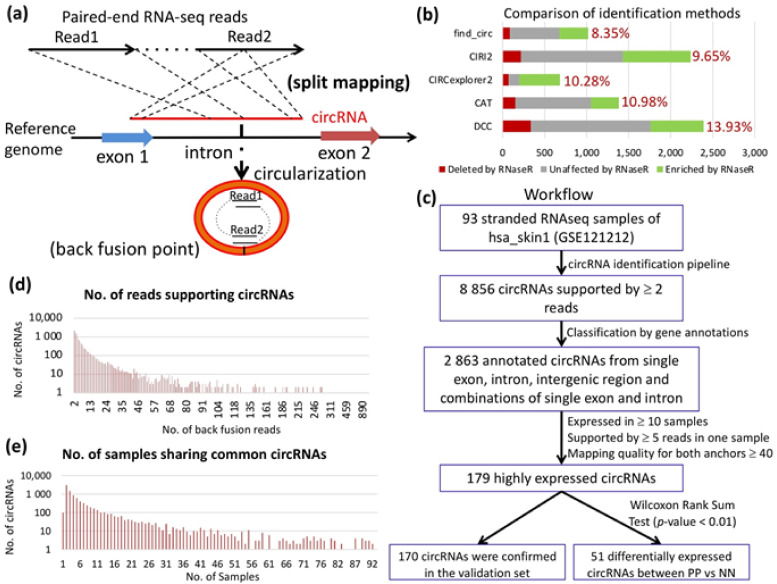
Figure 1.
Workflow of CAT and analysis of circRNAs in PS. (a) Illustration of split mapping of paired-end RNA-seq reads to candidate circRNAs. If Read1 is completely mapped and Read2 is split mapped to the reference genome, a circular structured RNA, circRNA, could be formed from this genomic locus with the back-fusion point identified from the split mapping. (b) Stacked bar plots of the circRNAs in HeLa cells (RNase R vs. control) that were predicted by five circular RNA identification methods, with results separated into RNase R resistant (≥ fivefold enrichment, green), unaffected (one to fivefold enrichment, gray) and RNase R sensitive (depleted in RNase R treated samples, red). Percentages refer to the fraction of RNase R sensitive circRNAs defined as false positives. The minimal number of supporting reads was set to 3 for all five methods compared. (c) Workflow for identification and analysis of circRNAs in the ribo-zero, stranded paired-end RNA-seq data from 93 PS samples. In total, 8856 unique circRNAs were identified and 2863 circRNAs were annotated and classified by single exons, single introns, pairs of adjacent exons and introns, and intergenic non-coding transcripts. A total of 179 highly expressed circRNAs were selected by three criteria, i.e., expressed in more than 10 samples, supported by more than five reads in at least one sample and filtered based on a mapping quality threshold of 40 for both anchors. This resulted in 51 differentially expressed circRNAs when PP vs. NN skin was compared by Wilcoxon rank sum test. (d) The number of BF reads supporting 8856 unique circRNAs, and (e) the number of samples expressing 8856 unique circRNAs, where the y-axes are logarithmic (log10).