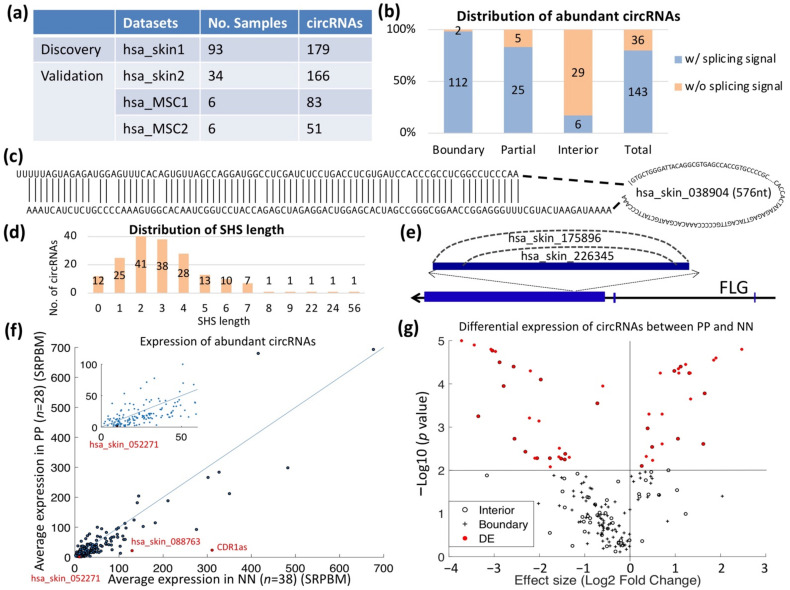
Figure 2.
Distribution and characteristics of abundant circRNAs. (a) Names and number of samples in the discovery and validation datasets. In total, 179 abundant circRNAs were identified in the discovery dataset, among which 166, 83, and 51 were independently validated by the three validation datasets. (b) Distribution of the 179 circRNAs in the discovery datasets. Boundary circRNAs are canonical circRNAs, and i-circRNAs are further classified into partial and complete i-circRNAs. All circRNAs are further annotated as to whether they are adjacent to splicing signals. (c) An example of RNA folding structure and complementary sequences flanking an exon i-circRNA, hsa_skin_038904. (d) Distribution of the lengths of short homologous sequences (SHS). (e) An example of two circRNAs, hsa_skin_175896 and hsa_skin_226345, arising from FLG (filaggrin). (f) Average expression (spliced reads per billion mappings [SRPBM]; see Methods) of the 179 highly expressed circRNAs in PP and NN skin. (g) Volcano plot visualizing differential expression of circRNAs when PP and NN skin samples were compared. The red and black dots in the plot represent significantly differentially expressed (p-value < 0.01, see Methods) and not significantly expressed circRNAs, respectively. Circle and cross marks represent interior and boundary circRNAs, respectively.