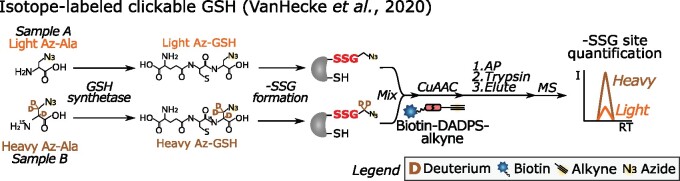
Figure 6.
Metabolic incorporation of isotopically labeled Az-Ala in GSH for site-specific quantification of –SSG (VanHecke et al., 2020). Light (orange) or heavy (brown) Az-Ala is administered to a GSH synthetase mutant cell line to metabolically produce Az-GSH. After protein extraction of mock- or H2O2-treated cells, the isotopically labeled Az –SSG sites (red bold font) are conjugated with biotin-dialkoxydiphenylsilane (DADPS)-alkyne, where the DADPS is acid sensitive. After streptavidin capture and on-bead trypsin digestion, isotope-labeled –SSG peptides are released by acidic cleavage of DADPS. MS analysis not only identifies the –SSG sites but also determines the relative –SSG site levels between samples by comparison of light- and heavy-labeled –SSG peptide intensities. I, intensity; –SSG, S-glutathione; RT, retention time.