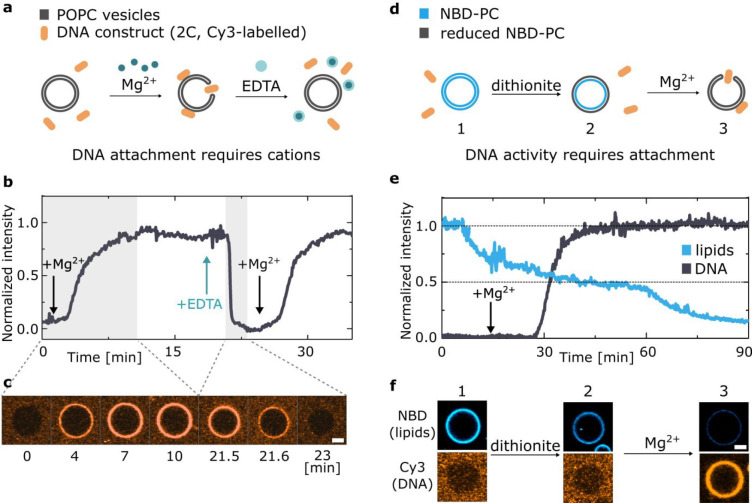
Figure 4.
Cation-regulated reversible DNA–membrane binding and activation of a synthetic enzyme. (a) Schematic representation of the mechanism leading to reversible DNA–membrane attachment upon addition of magnesium and its removal by means of chelating agent EDTA. (b) Representative fluorescence intensity trace of Cy3-labeled DNA nanostructures (2C) as recorded from POPC GUVs. DNA attachment and detachment are triggered by the addition of magnesium chloride and EDTA, respectively, as indicated by arrows. Delays associated with the diffusion of added Mg2+ and EDTA through the experimental chamber result in short lag times before changes in fluorescence are observed (for details see Section S3). (c) Confocal micrographs from the highlighted gray areas of the trace in (b), demonstrating the attachment and detachment transients. Scale bar: 5 μm. (d) Schematics of the NBD–dithionite reduction assay used to demonstrate cation-activated lipid scrambling. Upon addition of dithionite only the outer leaflet of NBD-tagged membrane is bleached. Magnesium addition induces the insertion of 2C DNA, which creates toroidal membrane pore and induces interleaflet mixing, leading to further fluorescence decrease. (e) Representative trace of the fluorescent intensity of NBD-labeled lipids (blue) upon addition of dithionite, alongside the trace representing Cy3-labeled DNA coating of the vesicle (black) appearing after addition of magnesium (arrow). See Figure S19 for a noninserting control and Figure S20 for an additional example. (f) Representative confocal micrographs, showing the fluorescence of both DNA and lipids at each stage of the experiment described in (d). Scale bar: 5 μm.