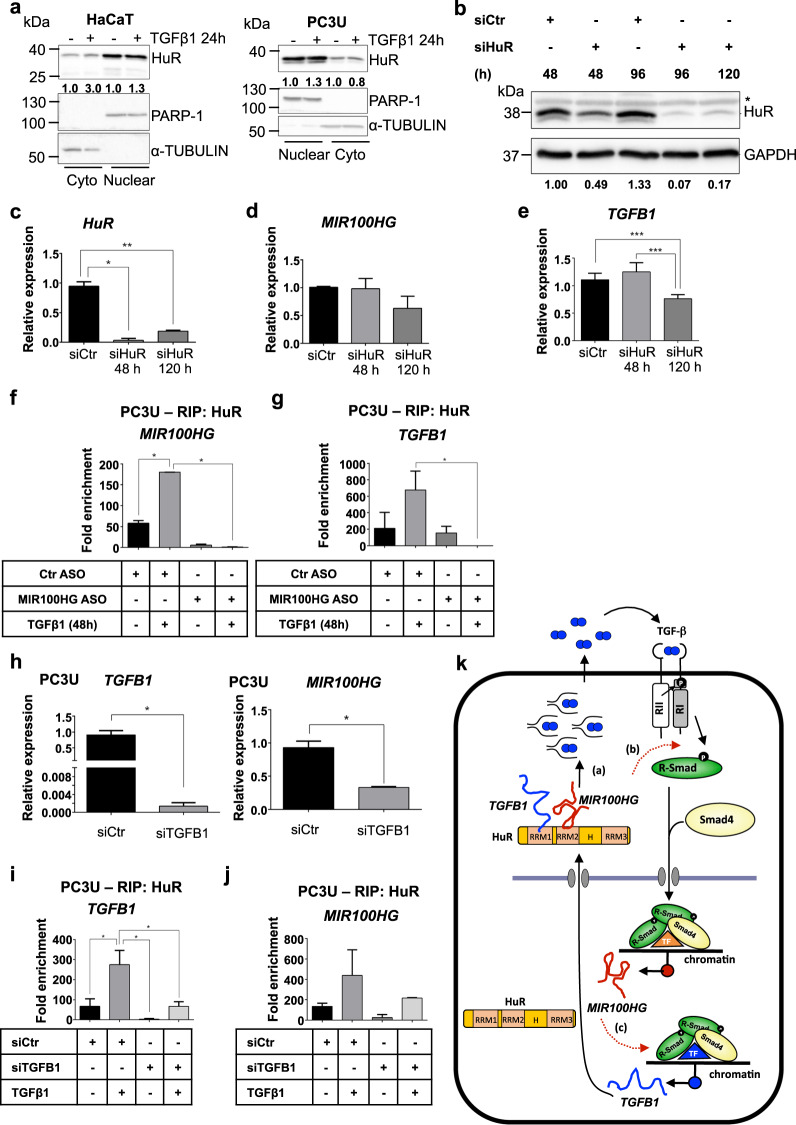
Fig. 7. MIR100HG engages HuR to regulate TGFβ1.
a Representative immunoblots out of two independent experiments for expression of HuR in HaCaT (left) and PC3U (right) cells treated with TGFβ1 for 24 h. PARP-1 (nuclear) and α-TUBULIN (cytoplasmic) were used as fractionation controls and molecular mass (kDa) markers are indicated along with densitometric values of normalized band intensity. b–e Representative immunoblot (b) out of three independent experiments for expression of HuR, and corresponding real-time RT-qPCR for detection of HuR (c), MIR100HG (d), and TGFB1 (e) mRNA expression in PC3U cells transiently transfected with negative control or specific siRNA for the indicated time periods. GAPDH was used as loading control and molecular mass (kDa) markers are indicated. A star indicates nonspecific protein bands recognized by the antibody. Gene expression is normalized relative to the housekeeping gene HPRT1. Error bars represent standard deviation from three different experiments (*P < 0.05, **P < 0.01, ***P < 0.001). f, g RIP analysis in PC3U cells transiently transfected with negative control or MIR100HG-specific ASO and stimulated with TGFβ1 or not for 48 h. Fold enrichment of the HuR-specific RIP relative to the IgG control is reported for MIR100HG (f) and TGFB1 (g) RNAs. Error bars represent standard deviation from three different experiments (*P < 0.05). h Real-time RT-qPCR for detection of TGFB1 and MIR100HG expression in PC3U cells transiently transfected with negative control or TGFB1-specific siRNA and treated with TGFβ1 for 48 h. Gene expression is normalized relative to the housekeeping gene HPRT1. Error bars represent standard deviation from three different experiments (*P < 0.05). i, j RIP analysis in PC3U cells transiently transfected with negative control or TGFB1-specific siRNA and stimulated with TGFβ1 or not for 48 h. Fold enrichment of the HuR-specific RIP relative to the IgG control is reported for TGFB1 (i) and MIR100HG (j) RNAs. Error bars represent standard deviation from three different experiments (*P < 0.05). k Diagrammatic scheme of TGFβ signaling regulating the downstream target genes MIR100HG and TGFB1, mediated by the two TGFβ receptor kinases and SMAD complexes together with gene-specific transcription factors (TF, note color differentiation based on gene specificity), leading to transcriptional induction of MIR100HG and TGFB1. Two possible (dashed red arrows) and one confirmed mechanism of action of MIR100HG are illustrated: (a) cytoplasmic MIR100HG associates with HuR in the cytoplasm (HuR domains are highlighted) and causes stabilization and accumulation of TGFB1 mRNA, which leads to enhanced synthesis of latent and mature secreted TGFβ1 (dimeric circles—mature TGFβ1—with twinkled lines—N-terminal pro-domain) that further stimulates the pathway in an autocrine manner. The two RNAs are shown to interact with distinct HuR RRM domains (not proven) and for completion, nuclear HuR is also illustrated. (b) Cytoplasmic MIR100HG promotes or stabilizes TGFβ receptor-SMAD complexes that prolong signaling. (c) A transcriptional mechanism in which nuclear MIR100HG enhances SMAD-mediated transcription of the TGFB1 gene.