Figure 6. An unbiased RNA sequencing screen identifies UCHL1-HIF-1 axis as a part of Txnip C247S-mediated molecular mechanisms.
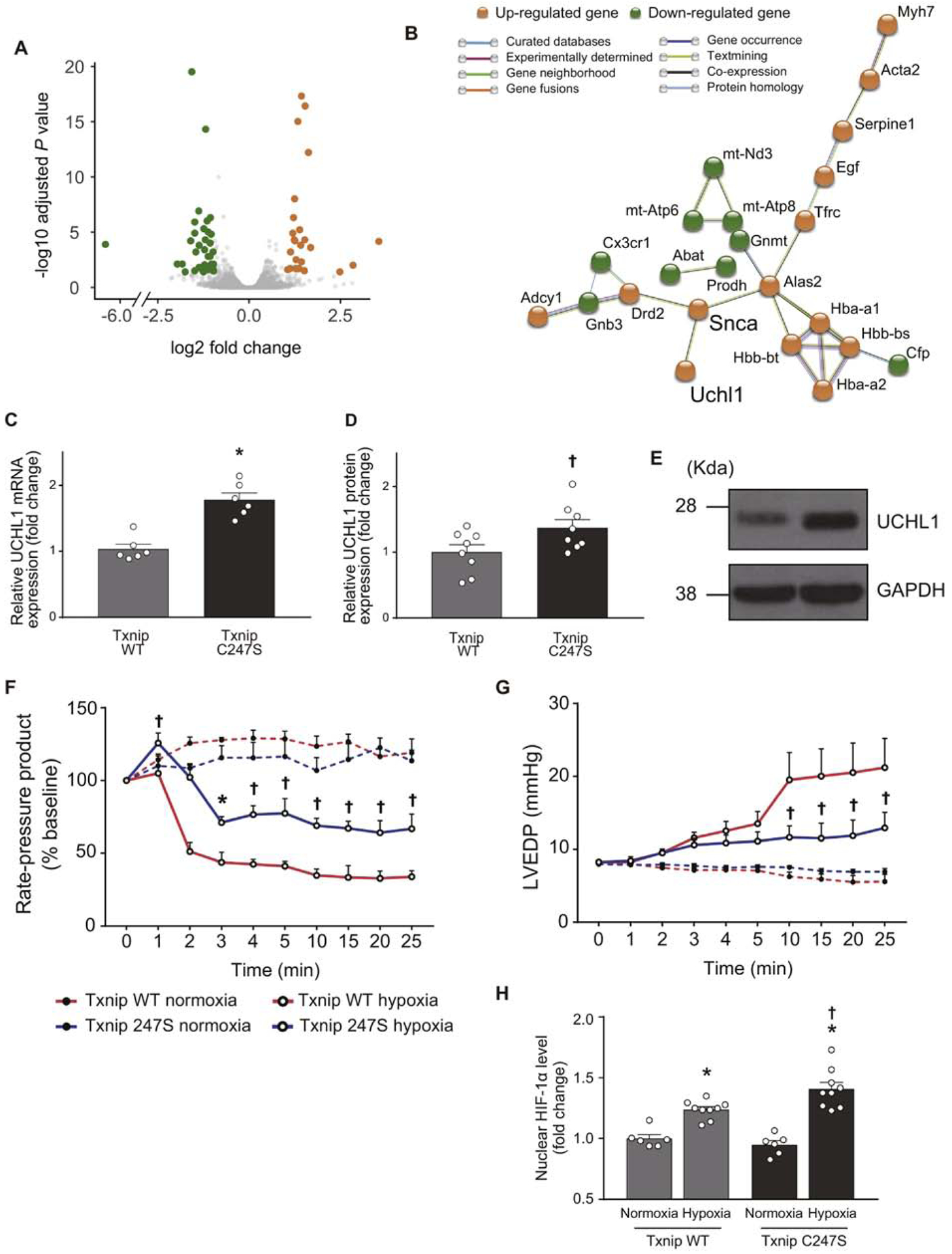
A. RNA-seq was performed in the heart tissues from Txnip wild-type (WT) and C247S mice (n=3 each). A volcano plot shows that 24 genes (orange) and 36 genes (green) were up- and down-regulated, respectively, in Txnip C247S hearts compared to control hearts. B. Functional enrichment analysis using STRING database identified a molecular interaction network mediated by Txnip C247S. UCHL1 is an activator of HIF1. C-E. Increased expression of UCHL1 were confirmed in Txnip C247S hearts by quantitative PCR (C) and Western blot analysis (D and E). *P<0.01 or †P<0.05 vs. WT. F-H. To test whether UCHL1-HIF-1 axis is functional, the Txnip C247S heart was subjected to global hypoxia (95% N2, 5% CO2; n=6) or normoxia (95% N2, 5% CO2; n=4) in Langendorff perfusion system. Txnip C247S hearts maintained better cardiac mechanical function during hypoxic perfusion, as shown by the higher rate-pressure product (F) and lower left ventricular end-diastolic pressure (LVEDP; G) than those in WT hearts. *P<0.05 or †P<0.01 vs. WT hypoxia. At the end of perfusion, HIF-1α levels were measured in nuclear extracts with ELISA. Txnip C247S hearts had higher HIF-1α levels to bind the HIF-1α response element under hypoxia (H). *P<0.01 vs. normoxia, and †P<0.05 vs. WT hypoxia.
