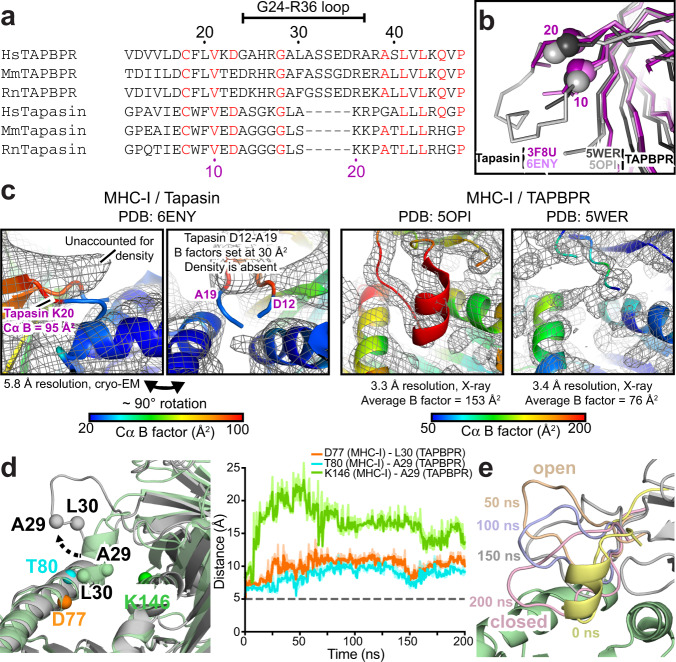
Fig. 1. Conformational plasticity of the TAPBPR G24-R36 loop.
a Sequence alignment between TAPBPR and tapasin from Homo sapiens (Hs, human), Mus musculus (Mm, mouse), and Rattus norvegicus (Rn, rat) highlighting differences in the TAPBPR G24-R36 loop region. Black and purple numbering reference TAPBPR and tapasin, respectively. Red residues are conserved. b Overlay of X-ray structures of tapasin (PDB IDs 3F8U and 6ENY) and TAPBPR (PDB IDs 5WER and 5OPI). c (Left) Cryo-EM density map (gray mesh, 5.0 σ) plotted on the cartoon of MHC-I in complex with tapasin (PDB ID 6ENY). (Right) 2Fo-Fc electron density maps (gray mesh, 1.0 σ) plotted on the cartoon of MHC-I in complex with TAPBPR (PDB ID 5OPI and 5WER). The cartoons are colored by Cα B factors. d (Left) Before (green) and during (gray) snapshots of peptide-deficient HLA-A*02:01/hβ2m/TAPBPR complex from all-atom MD simulations. The Cα atoms used for distance measurements are shown as spheres. (Right) Intermolecular Cα-Cα distances measured between HLA-A*02:01 groove (D77, T80, K146) and TAPBPR G24-R36 loop (A29, L30) residues over the course of the simulation. The dotted line represents Cα-Cα distances at the start of the simulation. e The range of conformations of the TAPBPR G24-R36 loop captured at different times during the MD simulation. The open (wheat, 50 ns) and closed (pink, 200 ns) states of the TAPBPR G24-R36 loop are oriented away from and covering the HLA-A*02:01 groove, respectively. The MHC-I groove (green) and TAPBPR N domain (gray) are shown as a static snapshot from the 0 ns MD time point for clarity.