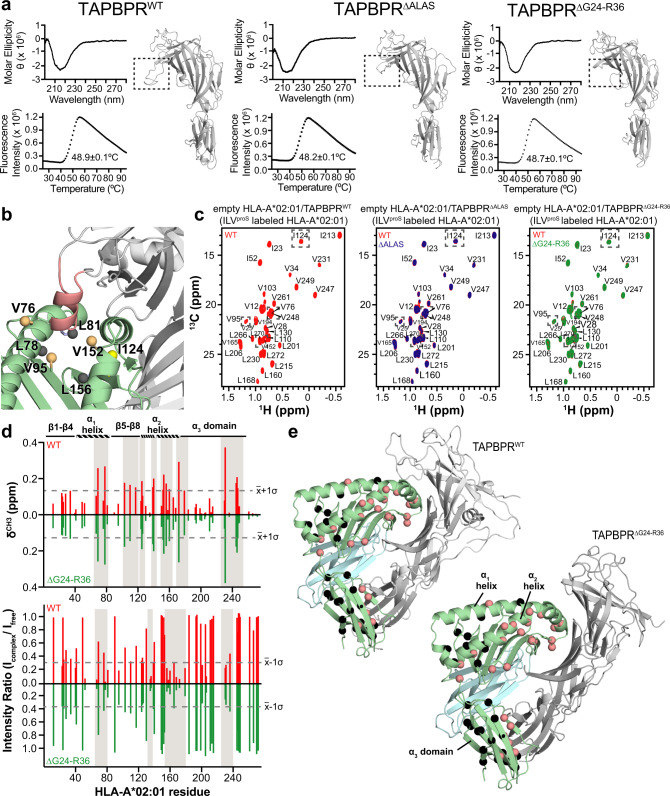
Fig. 4. The TAPBPR G24-R36 loop does not enter the HLA-A*02:01 groove.
a Comparison between wild-type (WT) and mutant TAPBPR constructs used in this study. ∆ALAS = deletion of residues A29-S32 and ∆G24-R36 = deletion of residues G24-R36. (Top) Far-UV CD and (Bottom) DSF spectra of each TAPBPR construct. Data are mean for n = 3 technical replicates. An inset in the DSF spectra notes measured thermal melt (Tm) values. The corresponding RosettaCM model of each TAPBPR construct is shown. The dotted box highlights differences in the G24-R36 loop region. b View of the peptide-deficient HLA-A*02:01/TAPBPR model (template PDB ID 5OPI) showing ILVproS methyl probes on HLA-A*02:01 (as spheres) within 10 Å of the TAPBPR G24-R36 loop (salmon). Methyl resonances of residues L78, L81 and L156 (shown in black) are missing in 2D 1H-13C methyl HMQC spectra of peptide-deficient HLA-A*02:01/TAPBPR complex due to conformational exchange induced line broadening. c 2D 1H-13C methyl HMQC spectra of 80 μM peptide-deficient HLA-A*02:01 (ILVproS labeled)/hβ2m in complex with TAPBPRWT (red), TAPBPR∆ALAS (blue) or TAPBPR∆G24-R36 (green) recorded at 25 °C at a 1H field of 800 MHz. Dotted boxes highlight methyl probes (shown in panel (b)) that are modeled to be near the TAPBPR G24-R36 loop. As a reference, the 2D 1H-13C methyl HMQC spectra of the complex formed with TAPBPRWT (red) is superimposed on those formed with TAPBPR∆ALAS or TAPBPR∆G24-R36. d Comparison of chemical shift deviations (CSD, δCH3, ppm) (top) and intensity changes (Icomplex/Ifree) (bottom) for AILV methyl probes of peptide-deficient HLA-A*02:01 in complex with either TAPBPRWT (red) or TAPBPR∆G24-R36 (green), relative to unbound TAX9/HLA-A*02:01. Dotted lines represent the average plus one standard deviation for CSD analysis (x̄ + 1σ) or minus one standard deviation for intensity ratio analysis (x̄-1σ). Gray boxes highlighted affected regions of HLA-A*02:01. The protein domains of HLA-A*02:01 are shown for reference. e Mapping of affected HLA-A*02:01 methyl probes exhibiting either CSD or intensity changes upon complex formation with TAPBPRWT or TAPBPR∆G24-R36 shown on RosettaCM models of the respective complex. Affected residues are colored salmon, unaffected residues are colored black.