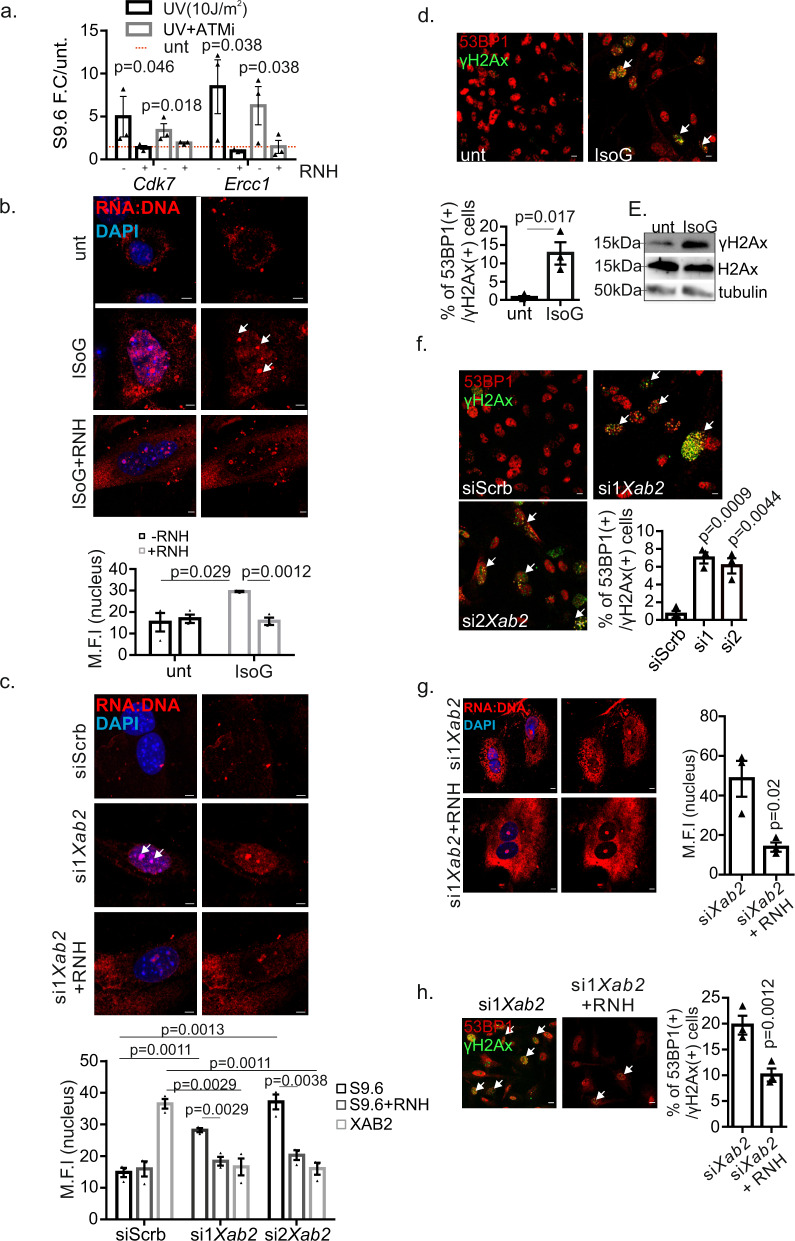
Fig. 6. Abrogation of XAB2 leads to R-loop-dependent genomic instability.
a DRIP analysis on promoter regions of Cdk7 and Ercc1 genes in untreated and UV-irradiated MEFs with or without RNase H1 (w/o RNH) and pretreated with ATM inhibitor (ATMi), as indicated. P values were calculated by one-tailed Student’s t test. DRIP signals are shown, as fold change over untreated. b–c Representative immunofluorescence images and quantification of RNA:DNA hybrids (indicated by the white arrows) in untreated and IsoG (IsoG)-treated wt. MEFs (see supplemental Fig. S4I for lower magnifications) and in siScrb and siXab2 MEFs, respectively (see supplemental Fig. S5A for lower magnifications and representative images for the efficiency of Xab2 silencing in si2Xab2 cells), w/o RNH. The graph depicts the mean S9.6 fluorescence intensity per nucleus (untreated ∓RNH: n = 1478/685 cells, IsoG treated ∓RNH: n = 1000/671cells, siScrb ∓ RNH: n = 1078/700 cells, si1Xab2 ∓RNH: n = 540/495 cells, si2Xab2 ∓ RNH: n = 640/580 cells/3 independent experiments)). Scale bars, 5 μm. d–f Immunofluorescence detection of γH2AX and 53BP1 (white arrowheads) in untreated and IsoG-treated wt. MEFs and in siScrb and siXab2 MEFs (see supplemental Fig. S5A for the efficiency of Xab2 silencing), respectively. The graph represents the percentage of γH2AX+ 53BP1+ cells (untreated: n = 1478 cells, IsoG-treated: n = 1045 cells, siScrb: n = 1095 cells, si1Xab2: n = 567 cells, si2Xab2: n = 723 cells/3 independent experiments)). Scale bar, 15 μm. e Western blot analysis of phosphorylated and total H2AX levels in untreated and IsoG-treated MEFs. The image shown is representative of experiments repeated three times. g Immunofluorescence detection of RNA:DNA hybrids in siXab2 MEFs with or without RNaseH1 (RNH) protein transfection. The graph represents the S9.6 fluorescence intensity per nucleus (siXab2∓RNH: n = 653/587 cells/3 independent experiments. Scale bar, 5 μm. h Immunofluorescence detection of γH2AX and 53BP1 (white arrowheads) in siXab2 MEFs with or without RNaseH1 (RNH) protein transfection. The graph represents the percentage of γH2AX+ 53BP1+ cells. (siXab2 ∓ RNH: n = 525/578 cells/3 independent experiments.).Scale bar, 15 μm. All scatter and bar blots are presented as mean ± SEM. P values were calculated by two-tailed Student’s t test, unless otherwise indicated. Error bars indicate SEM among three biological replicates. Source data provided as a Source Data file.