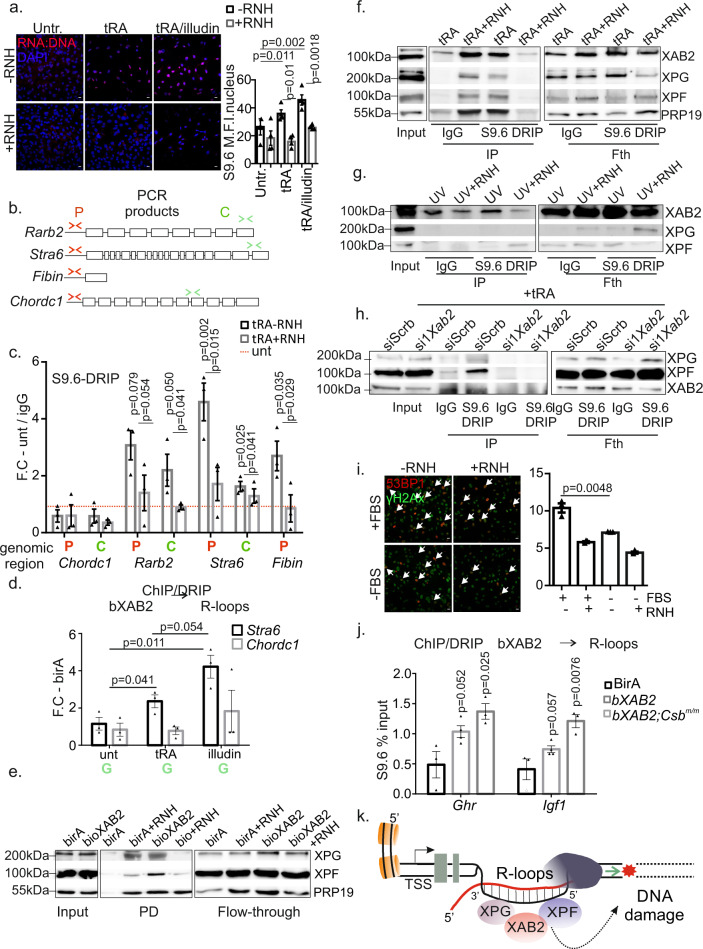
Fig. 7. High transcription promotes XAB2 interaction with R-loops.
a Immunofluorescence detection of R loops in untreated, tRA-treated (4 h) and illudin S-treated (1.5 h) MEFs (untreated ∓RNH: n = 975/705 cells, tRA treated ∓RNH: n = 1488/750 cells, illudin ∓RNH: n = 1500/730 cells/4 independent experiments.). b–c DRIP analysis of tRA-responsive (Rarb2, Fibin, and Stra6) and non-responsive (ChordC) gene promoters (P) or coding regions (c) w/o RNHin untreated and tRA-treated MEFs. p values between (−) and (+) RNH treated were calculated with one-tailed Student’s t test. d Native ChIP followed by DRIP on the tRA-induced Stra6 and the tRA-non induced Chordc1 coding regions in tRA and illudin S/tRA-treated BirA and bXAB2 MEFs. e bXAB2 pulldowns (PD) and western blotting for XPF, XPG, and PRP19 in tRA-treated bXAB2 and BirA MEF nuclear extracts w/o RNH. f–h S9.6 immunoprecipitation followed by WB for XAB2, XPG, XPF and PRP19 in (f) tRA-treated or (g) UV-irradiated (10 J/m2) MEF nuclear extracts w/o RNH and in (h) tRA-treated siScrb or si1Xab2 MEF nuclear extracts. i Immunofluorescence detection of γH2AX and 53BP1 (white arrowheads) in wt. MEFs cultured w/o fetal bovine serum (FBS) and transfected (RNH+) or not (RNH-) with RNaseH1. (+FBS ∓RNH: n = 1070/774 cells, −FBS ∓RNH: n = 785/831 cells/3 independent experiments) (j). Native ChIP followed by DRIP on the Ghr and Igf1 promoters in P15 wt. and Csbm/m livers. k DNA damage inhibits RNA synthesis. XAB2 is released from RNA targets leading to aberrant intron retention and R-loop accumulation. XAB2 interacts with ERCC1-XPF and XPG and the complex is recruited on RNA:DNA hybrids, linking R-loop processing with the spliceosomal response to DNA lesions. DRIP signals are shown, as fold enrichment (F.E) of % input of antibody over % input of control antibody (IgG). The images shown in Fig. 7e–h are representative of experiments that were repeated three times. All scatter and bar blots in this manuscript are presented as mean ± SEM. p values were calculated by two-tailed Student’s t test, unless otherwise indicated. Error bars indicate SEM among three biological replicates in all cases, unless otherwise stated. Source data are provided as a Source Data file.