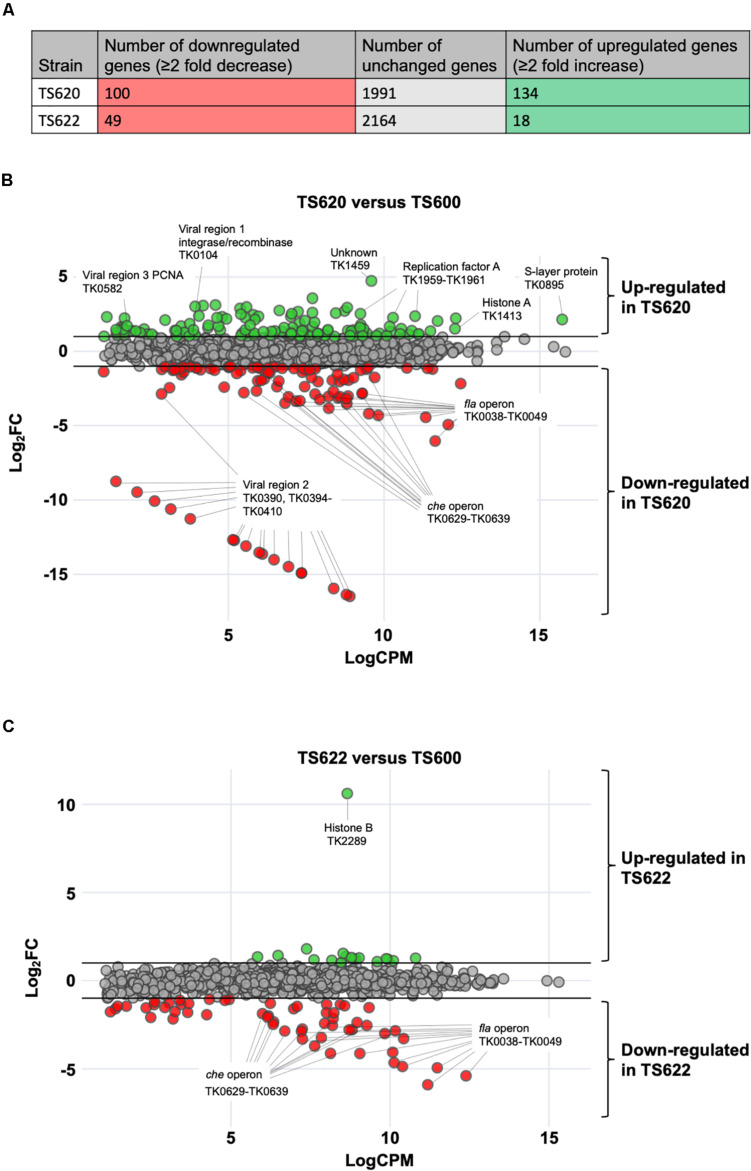
FIGURE 2.
Altering 3-dimensional chromatin structure dramatically alters gene expression in T. kodakarensis. (A) In TS620 (compared to TS600) 100 protein-coding genes were downregulated while 134 protein-coding genes were upregulated. In TS622 (compared to TS600), 49 protein-coding genes were downregulated while 18 protein-coding genes were upregulated. (B) In TS620 (TK1413G17D:histone A/ΔTK2289:histone B) differential RNA-sequencing, represented in an MA plot, revealed a number of genes are significantly upregulated and downregulated when compared to TS600 (TK1413WT:histone A/ΔTK2289:histone B). Green transcripts are significantly (≥2 fold change) enriched in TS620 when compared to TS600. Red transcripts are significantly depleted (≥2 fold change) in TS620 when compared to TS600. Notably, in TS620, a number of single stranded binding proteins (TK1959-1961: replication protein A subunits Rpa32, Rpa14, and Rpa41) are significantly enriched in TS620. TK1413G17D was found to be enriched in TS620 when compared to TK1413WT in TS600. In TS620 transcripts involved in cell motility and cell signaling (TK0038-TK0049: archaeal Fla operon and archaeal che operon (TK0629-TK0639) were significantly depleted. Additionally, a portion (TK0394-TK0410) of viral region 2 (TK0381-TK0421) was significantly depleted. (C) In TS622 (TK1413G17D:histone A/TK2289WT:histone B) far fewer genes were upregulated when compared to TS600. Downregulated genes related in cell motility and environmental signal sensing revealed similar trends to TS620 (che and fla operons).