FIGURE 2.
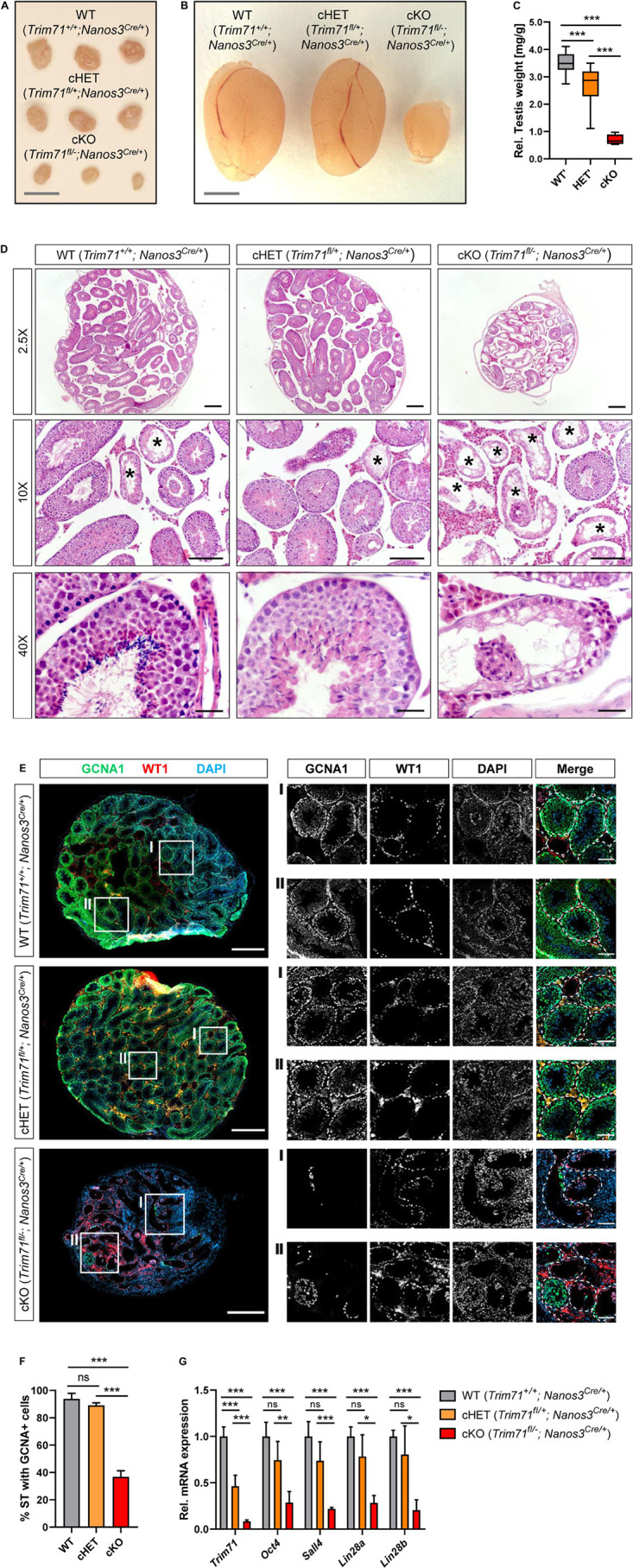
Germline-specific Trim71 cKO male mice present a Sertoli cell-only (SCO) phenotype. (A) Representative images of ovaries and (B) testes of adult wild type (WT, Trim71+/+; Nanos3Cre/+), germline-specific Trim71 heterozygous (cHET, Trim71fl/+; Nanos3Cre/+) and germline-specific Trim71 knockout (cKO, Trim71fl/−; Nanos3Cre/+) mice. Scale bars represent 2 mm. (C) Testis weight [mg] relative to total body weight [g] of wild type (WT’), Trim71 heterozygous (HET’) and germline-specific Trim71 knockout (cKO) mice, summarized from Supplementary Figure 2C, joining several Nanos3 genotypes per Trim71 genotype. Error bars represent SD (n = 8–29). ***P-value < 0.005 (one-way ANOVA, Tukey’s test). (D) Representative H&E stainings on paraffin testes cross-sections from adult wild type (WT, Trim71+/+; Nanos3Cre/+), germline-specific Trim71 heterozygous (cHET, Trim71fl/+; Nanos3Cre/+) and germline-specific Trim71 knockout (cKO, Trim71fl/−; Nanos3Cre/+) mice. Seminiferous tubules with a defective morphology are marked with an asterisk (*) in 10x magnification images. Scale bars represent 200, 100, and 20 μm in 2.5×, 10×, and 40× magnifications, respectively. (E) Representative immunofluorescence stainings on testes cryosections from adult wild type (WT, Trim71+/+; Nanos3Cre/+), germline-specific Trim71 heterozygous (cHET, Trim71fl/+; Nanos3Cre/+) and germline-specific Trim71 knockout (cKO, Trim71fl/−; Nanos3Cre/+) mice. Images show co-staining with GCNA1 (germ cells), WT1 (Sertoli cells) and DAPI (nuclei). For each genotype two regions – indicated as I and II – are depicted in higher magnification. Scale bars represent 500 μm for complete testes cross-sections and 100 μm for the magnification images. (F) Quantification of seminiferous tubules (ST) containing GCNA+ cells per testis cross-section from E, depicted as percentages. Error bars represent SD (n = 3). ***P-value < 0.005, ns, non-significant (unpaired Student’s t-test). (G) qRT-PCR of Trim71, the murine SSC markers Sall4, Lin28a and Oct4, and the differentiating spermatogonia marker Lin28b, relative to Hprt housekeeping gene in whole testis RNA of wild type (WT, Trim71+/+; Nanos3Cre/+), germline-specific Trim71 heterozygous (cHET, Trim71fl/+; Nanos3Cre/+) and germline-specific Trim71 knockout (cKO, Trim71fl/−; Nanos3Cre/+) male adult mice. Error bars represent SD (n = 3−6). ***P-value < 0.005; ***P-value < 0.01; *P-value < 0.05; ns, non-significant (unpaired Student’s t-test). See also Supplementary Figures 3, 4.
