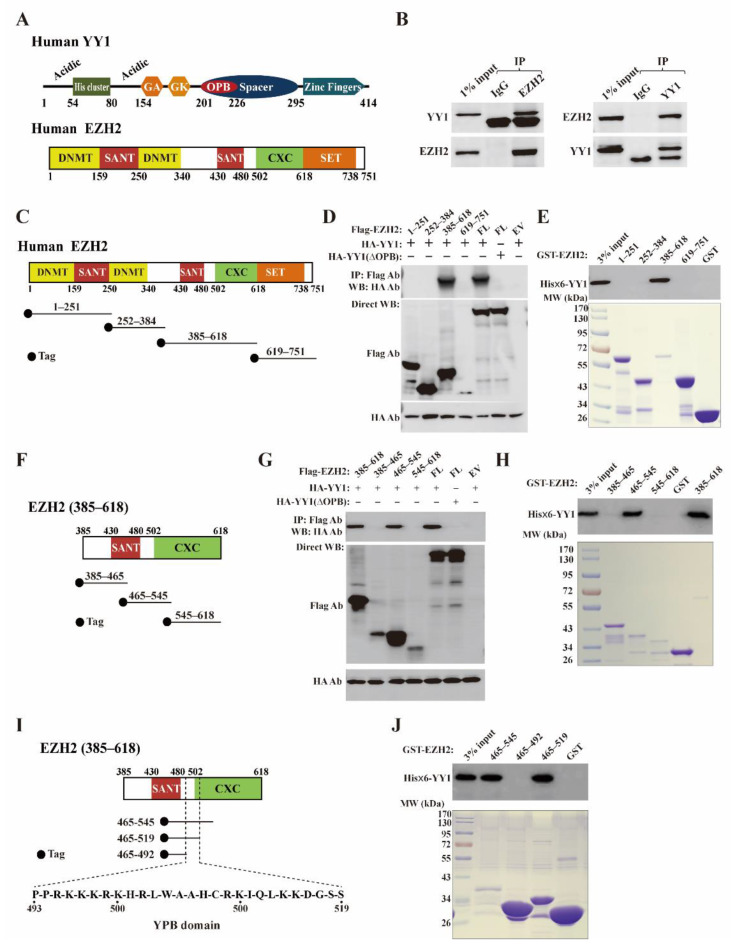
Figure 1.
Identification of YY1 binding site on EZH2 protein. (A) Schematic diagrams of the domain structures of YY1 and EZH2 proteins based on previous reports [30,39]. In the domain structure of YY1, “Acidic” represents a region containing enriched aspartic and glutamic acids, “His cluster” contains 11 consecutive histidines, “GA” and “GK” depict regions enriched by glycine/alanine and glycine/lysine, respectively; OPB: oncoprotein binding; the Spacer region and Zinc finger domain are also denoted. In the domain structure of EZH2, “DNMT” is the binding domain of DNA methyltransferase 1 (DNMT1), “SANT” denotes the SANT domain regulating chromatin accessibility [40],“CXC” is the cysteine-rich domain, and the SET domain is also denoted. (B) Western blot analyses of endogenous YY1 and EZH2 interaction by co-immunoprecipitation (co-IP). MDA-MB-231 cell lysates (800 µg) were incubated with 2 µg of EZH2 antibody (cat# 5246S, left) or YY1 antibody (cat# sc-281, right), followed by the incubation of Protein A/G beads and extensive washing. The samples were analyzed by Western blot using EZH2 and YY1 antibodies as indicated. (C,F,I) Diagram of EZH2 mutants generated to map its YY1 binding region. The tag is either an HA epitope or GST. (D,E,G–I) Co-IP (D,G) and GST-pull down (E,H,J) studies to determine the YY1 binding region on the EZH2 protein. FL: full length; EV: empty vector; IP: immunoprecipitation; Direct WB: Western blot analysis of samples without IP; YPB: YY1 protein binding.