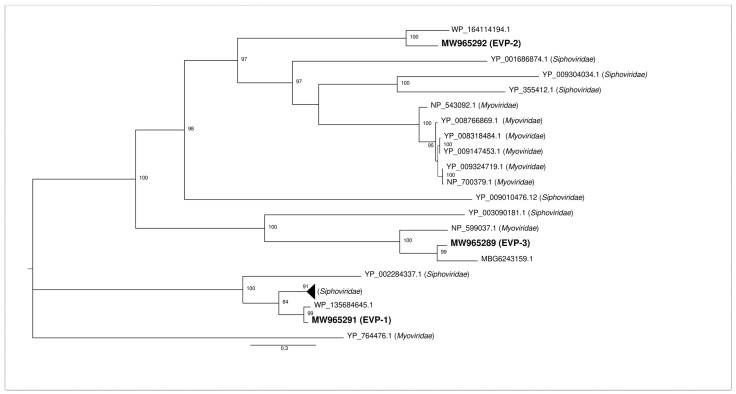
Figure 2.
Phylogenetic analysis of the deduced amino acid sequences of the three identified major capsid proteins. Major capsid proteins were aligned by MUSCLE and the phylogeny was inferred by the maximum-likelihood method using the CIPRES Science Gateway V. 3.3. Bootstrap analyses with 1000 iterations were used with the Dayhoff substitution matrix to estimate pairwise distances. Only bootstrap values higher than 70 are shown. The three identified major capsid proteins are in bold. As regards MW965289 and MW965291, which are both polycistronic transcripts, only the deduced amino acid sequences corresponding to the major capsid proteins were included in the alignment, i.e., ORF2 and ORF3, respectively. The scale bar indicates the evolutionary distance for 0.3 amino acid substitutions per site. The collapsed branch grouped nine sequences: BCI49740.1, Stx2a-converting phage Stx2 EH2201; BCI49860.1, Stx1a-converting phage Stx1 EH199; QIW91675.1, Escherichia phage Lys8385Vzw; YP_001449293.1, Enterobacteria phage BP-4795; YP_009909243.1 Enterobacteria phage 2851; YP_002274188.1, Enterobacteria phage YYZ-2008; BAT31949.1, Stx2-converting phage Stx2a F349; YP_009909397.1, Stx2-converting phage Stx2a WGPS6; and YP_009909320.1, Stx2-converting phage Stx2a WGPS8. GenBank accession numbers of the other major capsid proteins are: WP_135684645.1, Klebsiella pneumoniae; YP_002284337.1, Pseudomonas phage PAJU2; WP_164114194.1, Serratia marcescens; YP_009324719.1, Salmonella phage 118970 sal3; NP_700379.1, Salmonella phage ST64B; YP_008766869.1, Shigella phage SfIV; YP_009147453.1, Enterobacteria phage SfI; YP_008318484.1, Shigella phage SfII; NP_543092.1, Enterobacteria phage phiP27; YP_355412.1, Burkholderia phage Bcep176; YP_001686874.1, Azospirillum phage Cd; YP_009304034.1, Brucella phage BiPBO1; YP_009010476.1, Geobacillus phage GBK2; NP_599037.1, Enterobacteria phage SfV; YP_003090181.1, Burkholderia phage KS9; YP_764476.1, Geobacillus phage GBSV1; MBG6243159.1, Candidatus Symbiopectobacterium sp. Dall1.0.