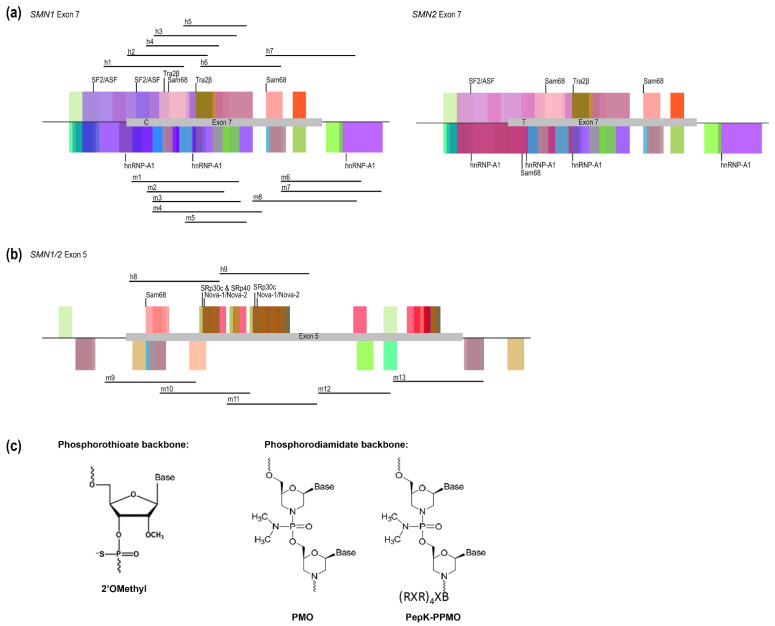
Figure 1.
(a) schematic of SMN1 and SMN2 exon 7 SpliceAid predicted splicing factor binding sites with the more strongly predicted splicing factors labelled, including alternative splicing factor/pre-mRNA splicing factor 2 (ASF/SF2) and transformer 2 Beta (Tra2β) as positive exonic splicing enhancer elements, and SRC associated in mitosis of 68 kDa (Sam68) and heterogenous nuclear ribonucleoprotein A1 (hnRNP-A1) as negative exonic splicing silencer elements. The binding location of the human exon 7 skipping AOs are indicated above the SMN1 exon 7, and the homologous mouse Smn exon 7 skipping AOs are indicated below; (b) schematic of SMN1/2 exon 5 predicted splicing factor binding sites, with the more strongly predicted splicing factors labelled, including Sam68, serine and arginine rich splicing factors (SRp-30c and SRp-40) and the Nova-1 and Nova-2 proteins as exonic splicing enhancers. The human and mouse exon 5 skipping AOs are indicated above and below the schematic, respectively; and (c) the chemical structure of the three AO chemistries used in this study, including the 2′-O-methyl phosphorothioate AO, the PMO and the PepK-conjugated PPMO, adapted from [27,28].