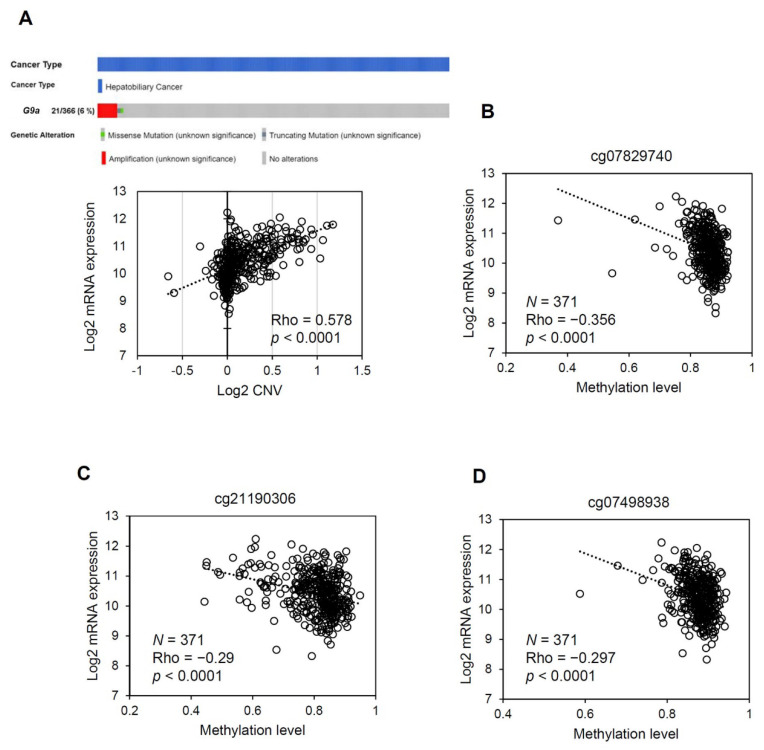
Figure 4.
G9a expression is controlled at both the genetic and epigenetic levels. (A) Upper panel: OncoPrint of G9a gene alterations in TCGA HCC dataset obtained from the cBio Cancer Genomics Portal (http://cbioportal.org, accessed on 25 January 2020). Lower panel: Copy number variants (CNVs) affect G9a gene expression. Positive and negative values respectively indicate gain and loss of the copy number. (B–D) Correlations of indicated CpG sites and G9a mRNA expression from TCGA HCC dataset (n = 371). Correlations were analyzed using the Spearman rank method. Regression lines, Spearman’s rho, and p-values are shown on the plot. Data used for correlation analyses were obtained via the Xena platform (https://xena.ucsc.edu/cite-us/, accessed on 25 January 2020).