Significance
RAS proteins (HRAS, NRAS, and KRAS) integrate extracellular trophic signals to promote cell proliferation. Constitutively active KRAS drives tumor initiation and progression in nonsmall cell lung cancer (NSCLC). As RAS proteins are often refractory to direct pharmacological inhibition, RAS-interacting proteins are under investigation as potential drug targets. We previously found Argonaute 2 (AGO2) to bind RAS and positively regulate RAS-dependent signaling pathways. AGO2 knockdown blunted proliferation in NSCLC cells containing a KRAS-activating mutation. Here, we demonstrate that AGO2 promotes tumor progression in multiple mouse models of KRAS-driven NSCLC. In these animals, Ago2 knockout impairs tumor growth, lowers pathologic grade, and inhibits KRAS signaling. Targeting the AGO2-KRAS interaction may hold future therapeutic promise in NSCLC and other KRAS-driven malignancies.
Keywords: AGO2, KRAS, nonsmall cell lung cancer
Abstract
Lung cancer is the deadliest malignancy in the United States. Non–small cell lung cancer (NSCLC) accounts for 85% of cases and is frequently driven by activating mutations in the gene encoding the KRAS GTPase (e.g., KRASG12D). Our previous work demonstrated that Argonaute 2 (AGO2)—a component of the RNA-induced silencing complex (RISC)—physically interacts with RAS and promotes its downstream signaling. We therefore hypothesized that AGO2 could promote KRASG12D-dependent NSCLC in vivo. To test the hypothesis, we evaluated the impact of Ago2 knockout in the KPC (LSL-Kras G12D/+;p53f/f;Cre) mouse model of NSCLC. In KPC mice, intratracheal delivery of adenoviral Cre drives lung-specific expression of a stop-floxed KRASG12D allele and biallelic ablation of p53. Simultaneous biallelic ablation of floxed Ago2 inhibited KPC lung nodule growth while reducing proliferative index and improving pathological grade. We next applied the KPHetC model, in which the Clara cell–specific CCSP-driven Cre activates KRASG12D and ablates a single p53 allele. In these mice, Ago2 ablation also reduced tumor size and grade. In both models, Ago2 knockout inhibited ERK phosphorylation (pERK) in tumor cells, indicating impaired KRAS signaling. RNA sequencing (RNA-seq) of KPC nodules and nodule-derived organoids demonstrated impaired canonical KRAS signaling with Ago2 ablation. Strikingly, accumulation of pERK in KPC organoids depended on physical interaction of AGO2 and KRAS. Taken together, our data demonstrate a pathogenic role for AGO2 in KRAS-dependent NSCLC. Given the prevalence of this malignancy and current difficulties in therapeutically targeting KRAS signaling, our work may have future translational relevance.
Lung cancer is the deadliest malignancy in the United States, causing more than a quarter of all annual cancer fatalities (1). Non–small cell lung cancer (NSCLC) accounts for 85% of lung cancer cases and is frequently driven by KRAS mutations (2). The RAS family, which also includes HRAS and NRAS, encodes small GTPases that transduce extracellular mitogenic signals by cycling between an active GTP-bound state and an inactive GDP-bound state. Intrinsic GTP hydrolysis is compromised in oncogenic RAS variants (3, 4). Resultant constitutive elevation of GTP-bound RAS promotes neoplastic transformation by hyperactivating downstream signaling pathways such as RAF-MEK-ERK (MAPK) and PI3 kinase (PI3K) (4–6). In NSCLC, activating RAS mutants encode oncogenic KRASG12C, KRASG12V, and KRASG12D (7). Efforts to design viable KRAS inhibitors have been ongoing for the last four decades, but hindered by key features of RAS structure and biochemistry. Structurally, RAS lacks adequate binding pockets for small molecule inhibitors. Biochemically, the active site binds GTP at picomolar affinity, hampering competitive inhibition (8, 9). Novel KRASG12C inhibitors have recently overcome these obstacles, showing clinical promise in treating solid tumors, including NSCLC (10, 11). Nonetheless, other oncogenic KRAS variants remain refractory. As such, recent work has aimed to identify KRAS-interacting proteins that may be targeted to suppress its downstream signaling in tumor cells.
Our group employed immunoprecipitation and mass spectrometry to screen KRAS-interacting proteins. We identified Argonaute 2 (AGO2)—a key regulator of microRNA (miRNA)-induced gene silencing—as a direct KRAS binding partner. AGO2-KRAS interaction occurred in NIH 3T3 cells ectopically overexpressing wild-type (WT) KRAS, and in several cancer lines harboring mutant KRAS variants. In KRASG12C-expressing H358 NSCLC cells, AGO2 knockdown inhibited cell proliferation, blunted colony formation, and reduced phosphorylation of RAS signaling mediators AKT, mTOR, and RSP6. Together, these findings suggested AGO2 could promote KRAS-driven malignancies through up-regulation of KRAS signaling (12). Indeed, a follow-up study demonstrated that AGO2 enables tumor progression in a mouse model of pancreatic ductal adenocarcinoma (PDAC) harboring KRASG12D (13).
AGO2 is one of four members of the mammalian Argonaute (AGO) family (which also includes AGO1, AGO3, and AGO4) (14, 15). AGO proteins are essential components of the RNA-induced silencing complex (RISC). Within RISC, miRNAs loaded onto AGO engage in complementary binding with target mRNAs to repress their translation (16, 17). AGO2 is unique among its family members in that it has intrinsic endonuclease activity and therein directly catalyzes the degradation of mRNA species (18). Moreover, AGO2 functions in DICER-independent miRNA processing to facilitate conversion of specific pre-miRNAs to mature miRNAs (19, 20). AGO2 also promotes miRNA stability, as demonstrated by experiments showing reduced miRNA half-life after AGO2 knockdown that can be restored with AGO2 reconstitution (21). Intriguingly, total miRNA levels are often lower in cancers than in the corresponding normal tissue (22). Moreover, reductions in specific miRNA species—including members of the let-7 family in NSCLC—have been directly linked to tumorigenesis (23, 24). In contrast, AGO2 is up-regulated in multiple tumor types (25–29); and data from The Cancer Genome Atlas (TCGA) show AGO2 gene amplification and increased AGO2 expression to be common in NSCLC (30). The discordance between expression patterns of miRNAs and AGO2 in clinical cancer samples suggests that AGO2 might promote tumorigenesis through miRNA-independent mechanisms, such as regulation of KRAS signaling (12). To date, however, loss-of-function studies examining the in vivo role of AGO2 in NSCLC have not been performed. Here, we employ multiple genetically engineered mouse models to test the hypothesis that AGO2 promotes KRAS signaling and drives tumorigenesis in NSCLC.
Results
Genetic Ablation of Ago2 Impairs Tumor Growth in Lung Adenocarcinoma Driven by KrasG12D Expression and p53 Loss.
In order to define the contribution of Ago2 to NSCLC, we analyzed the impact of its genetic ablation in the established KPC mouse model. KPC animals harbor both a stop-floxed KrasG12D transgene and two floxed p53 alleles. When subjected to intratracheal delivery of Cre recombinase-expressing adenovirus (Adeno-Cre) they develop tumors consistent with NSCLC (31, 32). By intercrossing KPC animals and mice harboring floxed Ago2 alleles (33), we generated progeny with concurrent homozygous (KPC-Ago2−/−) or heterozygous (KPC-Ago2+/−) ablation of Ago2. Sixteen weeks after administration of Adeno-Cre we analyzed tumor burden in these groups versus control KPC animals with wild-type Ago2 expression (KPC-Ago2+/+) (SI Appendix, Fig. S1A). Immunohistochemistry demonstrated robust Ago2 expression across the lung parenchyma and within tumor lesions of KPC-Ago2+/+ mice. Over 90% of lesions from lungs of KPC-Ago2−/− animals showed no detectable AGO2 staining, indicating good knockout efficiency (SI Appendix, Fig. S1 B and C).
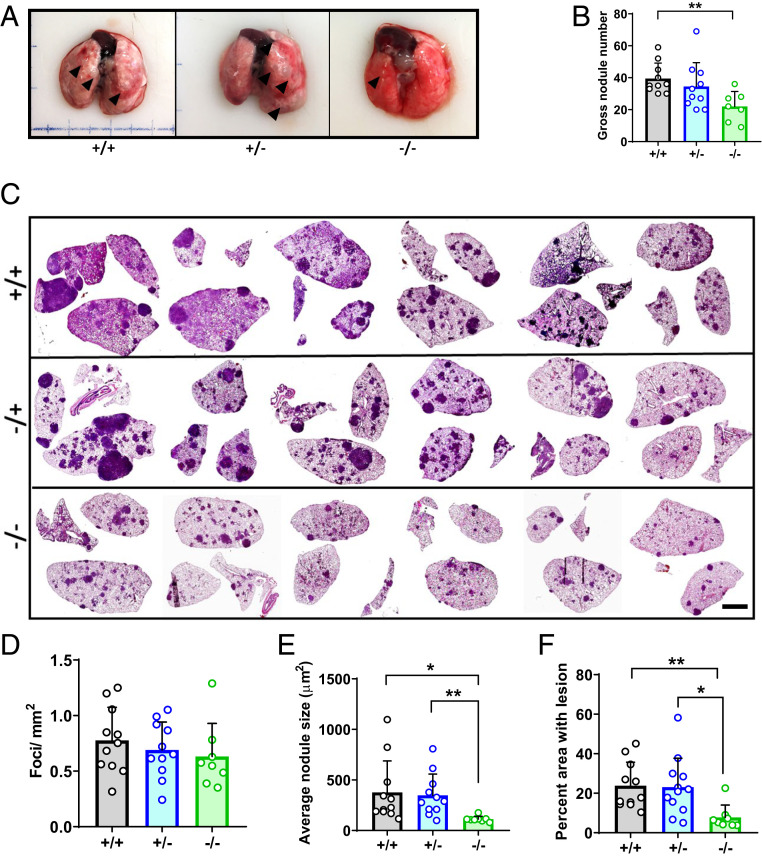
As previously reported (31) KPC-Ago2+/+ animals developed significant tumor burden at the 16-wk time point, with numerous nodules visible on gross inspection of the lungs (Fig. 1A). While the number of grossly visible nodules was unchanged in the KPC-Ago2+/− group, it was significantly lower in KPC-Ago2−/− mice versus KPC-Ago2+/+ controls (Fig. 1 A and B). Analysis of histological cross-sections, in which tumor inclusions are appreciable as hematoxylin-rich densities, showed that while nodule number per unit of cross-sectional area was unchanged among groups, average nodule size was markedly lower in KPC-Ago−/− animals versus both KPC-Ago2+/+ and KPC-Ago2+/− mice (Fig. 1 C–E). As such, the reduction in total lung area occupied by tumor tissue seen with homozygous Ago2 ablation (Fig. 1F) was driven by reduction in nodule size, rather than nodule number. Together, these findings indicate that AGO2 promotes NSCLC tumor growth.
Fig. 1.
Ago2 ablation impairs tumor growth in KPC lung cancer. (A) Gross lung samples from KPC-Ago2+/+ (+/+), KPC-Ago2+/− (+/−), and KPC-Ago2−/− (−/−) mice. Representative images from n = 11, n = 11, and n = 8 animals per group, respectively. Arrowheads indicate grossly visible tumor nodules. (B) Quantification of grossly visible nodules from aforementioned animals expressed as number of total nodules per pair of lungs. (C) Representative H&E-stained image series of lung cross-sections from +/+, +/−, and −/− mice. (Scale bar, 2.5 mm.) (D) Quantification of tumor foci per mm2 cross-sectional area. Average of three nonconsecutive sections from each of 11 +/+, 11 +/−, and 8 −/− animals. (E) Average surface area of individual nodules described in D. (F) Percentage of total lung cross-sectional area occupied by tumor lesion in the same animals as described in C–E. *P < 0.05, **P < 0.01.
AGO2 Facilitates Progression of KPC Lung Adenocarcinoma.
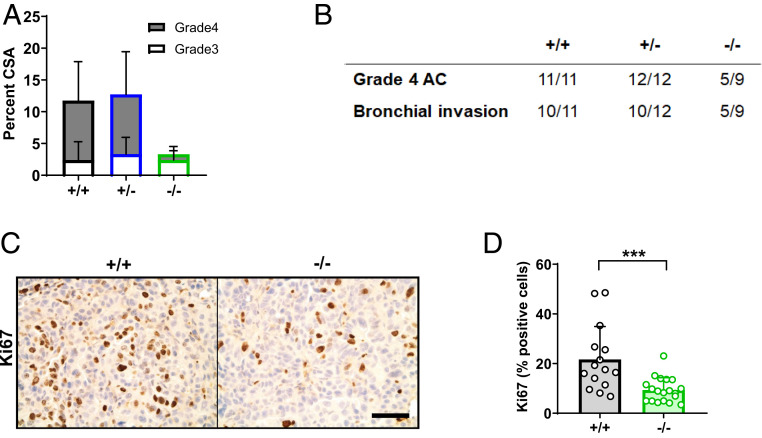
Adenocarcinoma comprises much of the lung tumor burden in the KPC model (31). In keeping with these findings, we observed advanced (grades 3 and 4) adenocarcinoma lesions (SI Appendix, Fig. S2A) within lungs of KPC mice 16 wk after Cre administration. Indeed, in KPC-Ago2+/+ and KPC-Ago2+/− mice, adenocarcinoma occupied >10% of total lung cross-sectional area (Fig. 2A). In contrast, adenocarcinoma occupied <3% of cross-sectional area in KPC-Ago2−/− animals (Fig. 2A). Furthermore, grade 4 adenocarcinoma comprised >75% of tumor lesion cross-sectional in KPC-Ago2+/+ and KPC-Ago2+/− mice, but only 25% in KPCAgo2−/− mice (Fig. 2A). Consistent with this, while grade 4 lesions, were uniformly observable in both KPCAgo2+/+ and KPCAgo2+/− animals, nearly half of KPCAgo2−/− mice were completely free of tumors reaching that pathologic grade (Fig. 2B). Notably, in KPCAgo2−/− mice, the infrequently observed AGO2(+) tumor nodules (SI Appendix, Fig. S1C) actually comprised the majority of grades 3 and 4 disease (SI Appendix, Fig. S2B). Indeed, AGO2(+) nodules within KPCAgo2−/− lungs had similar adenocarcinoma frequency to those in KPC-Ago2+/+ mice (SI Appendix, Fig. S2 B and C).
Fig. 2.
Ago2 facilitates progression of KPC lung adenocarcinoma. (A) Percentage of lung cross-sectional area occupied by adenocarcinoma (subdivided into grade 3 and grade 4) in KPC-Ago2+/+ (+/+) (n = 11), KPC-Ago2+/− (+/−) (n = 12), and KPC-Ago2−/− (−/−) (n = 9) mice. Analyses based on pathological assessment of H&E staining of whole lung cross-sections. (B) Number of mice per group (represented as a fraction of total animals) with any evidence of grade 4 adenocarcinoma (AC) or bronchial invasion. (C and D) Ki67 staining in adenocarcinoma lesions from +/+ and f/f animals with quantification (n = 3 sections/animal, 6 animals/group). (Scale bar, 50 μm.) ***P < 0.001.
KPC-Ago2−/− mice were also partially protected from the overt bronchial invasion seen in the vast majority of KPC-Ago2+/+ and KPC-Ago2+/− animals (Fig. 2B). Tumor proliferation index, assessed by Ki67 staining, was also significantly reduced with genetic Ago2 ablation (Fig. 2 C and D). Finally, the reduced rate of tumor progression observed in the KPC-Ago2−/− group corresponded with increased survival versus animals of other genotypes (SI Appendix, Fig. S3).
Ago2 Ablation Also Lowers Tumor Burden in Clara Cell–Derived, KrasG12D-Dependent Lung Cancer.
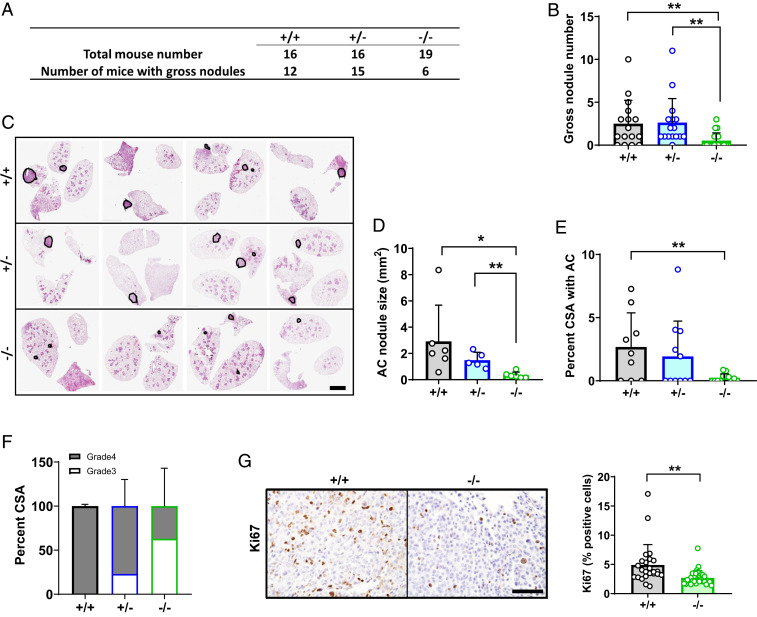
We next interrogated AGO2 function in two additional NSCLC models by employing animals that contain a tamoxifen (Tam)-inducible Cre recombinase gene driven by the (Clara cell–specific) CCSP (CC10) promoter (34). In the first model, we intercrossed Tam-inducible CCSP-Cre animals with stop-floxed KrasG12D mice to enable time-dependent activation of Kras oncogene. This is commonly referred to as the KC model (SI Appendix, Fig. S4A). As previously reported, KC mice develop epithelial hyperplasia by 3 wk, adenoma by 6 wk, and occasional adenocarcinoma 15 wk after Cre activation. The lungs of these animals also contain a macrophage-predominant inflammatory infiltrate (35). By intercrossing Ago2 floxed animals into the KC line, we generated KCAgo2+/+, KCAgo2+/−, and KCAgo2−/− mice and treated with Tam (50 mg/kg, delivered by intraperitoneal injection) at 6 wk of age to initiate tumor formation (SI Appendix, Fig. S4B). Sixteen weeks later, we killed the animals and noted lung nodules in KCAgo2+/+ mice, as previously described (35) (SI Appendix, Fig. S4C). Pathologic subtypes ranged from hyperplasia to frank adenocarcinoma, while inflammation was broadly appreciable in lungs of most animals (SI Appendix, Fig. S4D). Though the frequencies of hyperplasia and adenoma were largely unchanged with Ago2 ablation, adenocarcinoma was absent in all KCAgo2−/− mice analyzed (SI Appendix, Fig. S4D). Unfortunately, the prominent lung inflammation inherent in this model complicated analysis of AGO2 function, as several mice in all groups died prematurely from pulmonary edema.
To circumvent the interference of inflammation with tumor analysis, we generated a more aggressive NSCLC model by employing the inducible CCSP Cre to simultaneously ablate one p53 allele while activating KrasG12D transcription. These mice, here called KPHetC, also sequentially develop alveolar adenoma and adenocarcinoma (36). We interbred KPHetC animals with mice harboring floxed Ago2 alleles to obtain KPHetCAgo2+/+, KPHetCAgo2+/−, and KPHetCAgo2−/− progeny (SI Appendix, Fig. S5A). In each group, we induced Cre activation with Tam at 6 wk old, then evaluated tumor burden 18 wk later. Upon necropsy, gross nodules were visible in more than 75% of KPHetCAgo2+/+ and KPHetCAgo2+/− mice. On the contrary, only 6 of 19 KPHetCAgo2−/− mice had evidence of these tumors (Fig. 3A). Average number of gross nodules per animal was also lower in KPHetCAgo2−/− mice versus the other groups (Fig. 3B). In histological cross-sections, the size of individual adenocarcinoma lesions was lower in a KPHetCAgo2−/− versus the other genotypes, while total area occupied by adenocarcinoma was lower in KPHetCAgo2−/− than KPHetCAgo2+/+ mice (Fig. 3 C–E and SI Appendix, Fig. S5B). We further observed that adenocarcinoma lesions in lungs of KPHetCAgo2+/+ mice were uniformly grade 4. Less advanced grade 3 lesions were more common with loss of 1 or both copies of Ago2 (Fig. 3F). Reduction in tumor nodule size and pathologic grade corresponded with lower cell proliferation (as indicated by Ki67 index) in KPHetCAgo2−/− mice (Fig. 3G). Taken together, these data demonstrate that AGO2 is required for the growth and progression of Clara cell–derived NSCLC.
Fig. 3.
Ago2 ablation impairs tumor growth in KPHet-C lung cancer. (A and B) Quantification of grossly visible nodules from lungs of KPHetC-Ago2+/+ (+/+), KPHetC-Ago2+/− (+/−), and KPHetC-Ago2−/− (−/−) mice. (C) Representative H&E-stained image series of lung cross-sections from +/+, f/+, and f/f mice. (Scale bar, 2.5 mm.) (D) Quantification of adenocarcinoma (AC) foci per mm2 cross-sectional area. (E) Average lung cross-sectional surface area (CSA) occupied by adenocarcinoma. (F) Percent grade 3 and grade 4 tissue within adenocarcinoma lesions of mice of each genotype. (G) Ki67 staining (Left images) and proliferative index (Right) in +/+ and −/− mice. (Scale bar, 50 μm.) n = 16 to 19 mice/group. *P < 0.05, **P < 0.01.
Ago2 Ablation Suppresses KRAS Signaling in NSCLC.
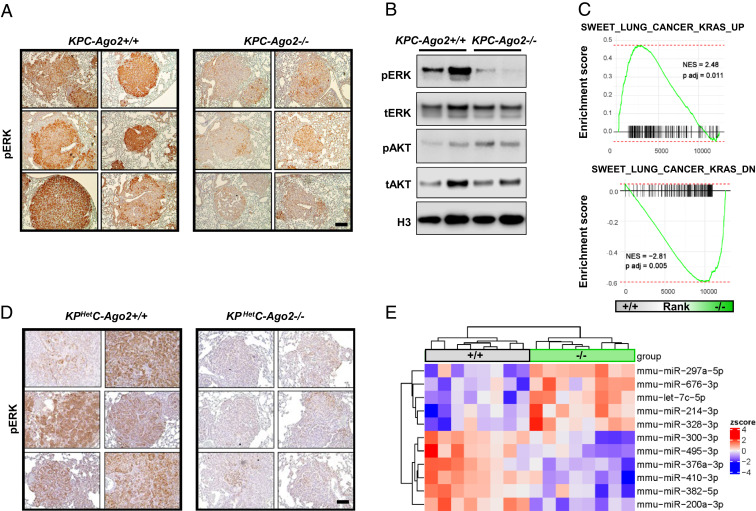
Since our previous work identified AGO2 as a positive regulator of canonical KRAS signaling (12), we performed immunohistochemistry for phosphorylated ERK (pERK) and noted reduced staining in lung nodules of KPC-Ago2−/− versus KPC-Ago2+/+ mice (Fig. 4A). Immunoblot similarly demonstrated lower pERK levels in the setting of Ago2 ablation (Fig. 4B). In contrast, immunoblots for phosphorylated AKT (pAKT) were similar between groups (Fig. 4B). Moreover, immunohistochemistry showed pAKT staining to be intense in ductal cells, but only scantly positive in tumor nodules, with no obvious intergroup differences (SI Appendix, Fig. S6A). To determine whether Ago2 ablation reduced KRAS-dependent gene expression in the same lesions, we subjected nodule isolates to RNA sequencing (RNA-seq), then performed gene set enrichment analysis (GSEA) employing signatures defined in KRAS-driven mouse lung cancer (37). Our analysis revealed KRAS-induced genes to be down-regulated and KRAS-repressed genes to be up-regulated in KPC-Ago2−/− versus KPC-Ago2+/+ animals (Fig. 4C). Together, these data indicate AGO2 positively regulates MAPK signaling in KPC lung cancer. Tumor nodules from KPHetC mice were too small to be individually isolated; however, analysis of pERK distribution by immunohistochemistry also revealed reduced staining in KPHetCAgo2−/− versus KPHetCAgo2+/+ nodules (Fig. 4D).
Fig. 4.
Ago2 ablation reduces Kras signaling in vivo. (A) pERK immunohistochemistry of lung cross-sections from KPC-Ago2+/+ versus KPC-Ago2−/− mice. (Scale bar, 50 μm.) (B) Immunoblot analysis of protein lysates from isolated lung nodules of KPC-Ago2+/+ and KPC-Ago2−/− mice. Phosphorylated ERK (pERK), total ERK (tERK), phosphorylated AKT (Ser-473) (pAKT), total AKT (tAKT), and histone H3 (H3). (C) GSEA assessing genes associated with increased KRAS signaling (SWEET_LUNG_CANCER_KRAS_UP and SWEET_LUNG_CANCER_KRAS_DOWN). Comparison of genes differentially expressed in KPC-Ago2+/+ versus KPC-Ago2−/− (isolated nodules). NES, normalized enrichment score. (D) pERK immunohistochemistry of lung cross-sections from KPHetC-Ago2+/+ and KPHetC-Ago2−/− mice. (Scale bar, 100 μm.) (E) Heat map of differentially expressed (false discovery rate [FDR] < 0.05) species from miRNA QPCR performed on nodules from KPC mice with normal Ago2 (+/+) or with biallelic Ago2 knockout (−/−).
We then applied high-throughput QPCR to assess levels of 239 miRNAs in tumor nodule samples isolated from KPC-Ago2+/+ and KPC-Ago2−/− mice. We found 11 species to be differentially expressed between groups (Fig. 4E). Among those up-regulated with Ago2 ablation was let-7c (Fig. 4E). This and other members of the let-7 family collectively decline in KRAS-driven NSCLC, promoting tumorigenesis (23, 24). Notably, MAPK signaling itself reduces let-7 levels by stimulating LIN28, an inhibitor of let-7 maturation (24). As such, impact on miRNA dynamics—either direct, or via canonical KRAS signaling—represents an interrelated mechanism by which AGO2 induces NSCLC progression in the KPC model. miR-214—a species for which both oncogenic and tumor suppressor roles have been reported in lung NSCLC (38–40)—was also elevated in KPC-Ago2−/− samples.
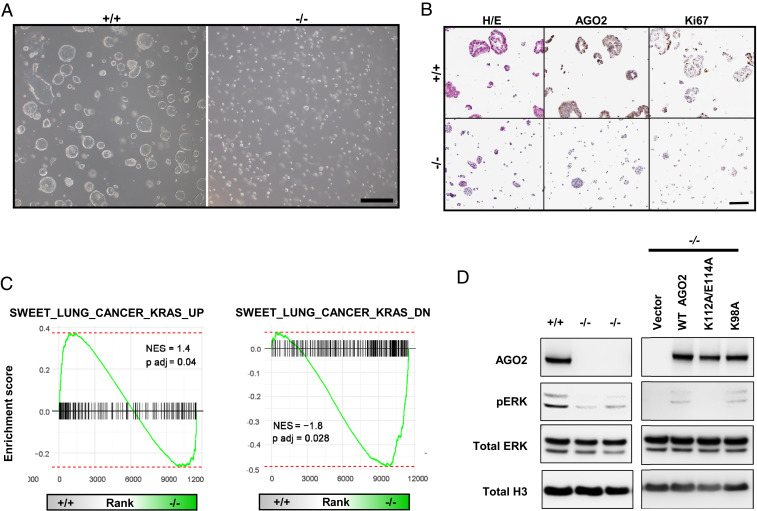
To further dissect the mechanistic link between AGO2 and canonical KRAS signaling in this system, we dissociated KPC tumor nodules and cultured cells ex vivo as organoids (41, 42). Consistent with our in vivo findings, organoids derived from KPC-Ago2−/− nodules were smaller (Fig. 5 A and B) and had lower proliferation indices (Fig. 5B) than KPC-Ago2+/+ organoids. RNA-seq with GSEA recapitulated findings from fresh nodule isolates, showing Ago2 ablation to confer significant reduction in KRAS-induced genes and increase in KRAS-repressed genes (Fig. 5C). We previously defined direct protein–protein interaction as required for AGO2 regulation of KRAS signaling (12). In cells derived from organoids and grown in 2D culture, immunoprecipitation confirmed AGO2-RAS binding (SI Appendix, Fig. S6B). Organoid-derived cells lacking Ago2 had lower proliferative rates (SI Appendix, Fig. S6 C and D) and pERK levels (Fig. 5 D) than those with intact Ago2. In order to determine whether direct interaction with KRAS was required for MAPK signaling in these cells, we transfected KPC-Ago2−/− cells with constructs encoding 1) wild-type AGO2, 2) AGO2K112A/E114A [deficient KRAS binding (12)], and 3) AGO2K98A [able to bind to KRAS (12)]. While add-back of WT AGO2 and AGOK98A both induced higher pERK levels, addition of AGO2K112A/E114A did not (Fig. 5 D). These findings demonstrate that in KPC tumor cells AGO2 promotes canonical MAPK signaling through direct interaction with AGO2.
Fig. 5.
AGO2 promotes MAPK signaling through direct interaction. (A) Organoids isolated from lung nodules of KPC-Ago2+/+ versus KPC-Ago2−/− mice. Bright field image. (Scale bar, 500 μm.) (B) Histological sections of KPC-Ago2+/+ and KPC-Ago2−/− organoids stained with H&E and antibodies directed against AGO2 and Ki67. (Scale bar, 150 μm.) (C) GSEA assessing genes associated with increased KRAS signaling (SWEET_LUNG_CANCER_KRAS_UP and SWEET_LUNG_CANCER_KRAS_DOWN). Comparison of genes differentially expressed in KPC-Ago2+/+ versus KPC-Ago2−/− organoids. NES, normalized enrichment score. (D, Left) Immunoblot analysis of protein lysates from KPC-Ago2+/+ versus KPC-Ago2−/− organoid-derived cells. Phosphorylated ERK (pERK), total ERK (tERK), and histone H3 (H3). (D, Right) Immunoblot analysis of KPC-Ago2−/− organoid-derived cells transfected with constructs encoding wild-type AGO2 (WT AGO2), AGO2K112A/E114A (K112A/E114A), and AGO2K98A (K98A).
Discussion
In Western countries, activating KRAS mutations underlie 30% of NSCLC and predict negative outcomes in early- and late-stage disease (43). The clinical relevance of aberrant RAS signaling in this and other cancer types has prompted a long-standing search for effective RAS inhibitors that has been hampered by structural and biochemical features of the RAS molecule (8, 9). Attempts to block downstream effectors, disrupt RAS subcellular localization, and identify synthetic lethal partners have also met with only limited clinical success (44). In this setting, the search for KRAS interaction partners represents an alternate approach for identifying new drug targets. We recently defined AGO2 as a binding partner of both native KRAS and its mutant variants. Interaction with AGO2 promotes KRAS downstream signaling, and AGO2 knockdown inhibits proliferation and colony formation in KRASG12C-expressing H358 NSCLC cells (12). In the current study, we establish AGO2 as an in vivo mediator of NSCLC progression and positive regulator of MAPK signaling through direct KRAS binding.
KRAS mutant tumors are relatively unique among oncogene-driven NSCLCs in that they often harbor additional genetic alterations, including mutations in STK11, CDKN2A/B, and p53 (45). Genetically engineered NSCLC mouse models recapitulate this aspect of human disease through conditional expression of one KrasG12D allele and ablation of one or both p53 alleles (32). In KPC mice, intratracheal Adeno-Cre administration yields KrasG12D expression and biallelic p53 knockout predominantly in alveolar type II cells (with lesser contribution from Clara cells). Early hyperplastic and adenomatous lesions precede the development of frank adenocarcinoma (46). Biallelic Ago2 ablation in KPC mice yielded smaller tumor nodules with reduced proliferative index and pathologic grade. In the KPHetC model, the Clara cell–specific CCSP Cre drives expression of KrasG12D allele and knockout of a single p53 allele. Our observation of inhibited tumor progression, reduced proliferative index, and down-regulated MAPK signaling in KPHetCAgo2−/− mice demonstrates that the effects of Ago2 ablation are consistent across mouse NSCLC models with different primary cells of origin and p53 loss status.
In both KPC and KPHetC models, inhibition of tumor progression corresponded with lower pERK levels in tumor nodules and/or nodule-derived organoids. Conversely, PI3K signaling was not obviously impacted. Our findings parallel those of several groups who demonstrated amplification of canonical MAPK signaling corresponds with progression of KRASG12D-driven murine adenocarcinoma (47–49). Clinical data demonstrating partial efficacy of the Mek1/2 inhibitor selumetinib in prolonging progression-free survival in KRAS-mutant NSCLC suggests a direct causative role for MAPK signaling in progression of analogous human tumors (50). Furthermore, Cicchini et al. directly tested this premise through forced amplification of MAPK signaling in nascent and established murine lung adenocarcinoma. In KRASG12D-expressing tumors derived from both alveolar type 2 cells and Clara cells, MAPK signaling was necessary for progression—particularly in p53-mutant disease (51). As such, our observation of reduced pERK in Ago2−/− KPC and KPHetC mice strongly suggests Ago2 ablation limits tumor progression by down-regulating MAPK signaling. The coincident increase of let-7c-5p observed in KPC-Ago2−/− versus KPC-Ago2+/+ nodules may also occur downstream of MAPK signaling and represents a parallel mechanism by which Ago2 loss impairs tumor progression in this model.
While our findings can be well integrated into the existing literature on KRASG12D-driven mouse models of lung cancer, comparison with other cell types in which AGO2 is genetically manipulated or inhibited reveals interesting differences that suggest context-specific consequences of KRAS-AGO2 interaction. For instance, in KRASG12C-expressing H358 cells AGO2 principally promotes RAS signaling through the PI3K pathway, as AGO2 knockdown causes a reduction in pAKT (12). In contrast, the KC mouse model of PDAC—in which the p48 Cre drives KrasG12D expression in the exocrine pancreas—undergoes bona fide oncogene-induced senescence upon genetic Ago2 ablation—blocking progression of early lesions to advanced adenocarcinoma (13). Interestingly, in that model, Ago2 ablation had no effect on PDAC progression in the setting of p53 loss (13). Taken together, these findings suggest AGO2 inhibition might be applied to target multiple forms of KRAS-driven cancer with different downstream effector pathways (44). Based on this potential pharmacologic flexibility, the AGO2-KRAS interaction warrants further study in NSCLC and other KRAS-driven malignancies.
Methods
Animals.
Mice of the following strains were obtained from The Jackson Laboratory: Ago2f/f [B6.129P2(129S4)-Ago2tm1.1Tara/J, 016520] (33), KrasLSL-G12D (B6.129S4-krastm4Tyi/J, 08179) (46), p53f/f (B6.129P2-Trp53tm1Brn/J, 008462) (52), and CCSP-Cre [B6N.129S6(Cg)-Scgb1a1tm1(cre/ERT)Blh/J] (34). Jackson breeds each on a C57BL/6J background. At our institution, animals were maintained in pathogen-free housing with a 12-h light/dark cycle and ad libitum access to food and water. Equal numbers of male and female mice were used for all experiments. The University of Michigan Institutional Animal Care and Use Committee approved all animal work.
Intratracheal Adenovirus Administration.
Cre-expressing adenovirus (Adeno-Cre) was obtained from the Viral Vector Core Facility at the University of Iowa. The virus was delivered by intratracheal administration as described (31). Briefly, mice aged 6 to 8 wk were anesthetized with isoflurane and then infected with 1.25 × 107 plaque-forming units (PFUs) of Adeno-Cre in 67.5 μL vehicle through a micropipette tip inserted into the pharynx. After mice regained consciousness, the same procedure was repeated, such that each mouse received a total of 2.5 × 107 PFUs in 125 μL vehicle. Viral administration was performed in a biosafety level 2+ room, in accordance with the guidelines of the University of Michigan Institutional Biosafety Committee.
Tamoxifen Administration.
Tamoxifen (Sigma) was dissolved in corn oil to a concentration of 20 mg/mL and administered to 6-wk-old mice by intraperitoneal injection at a dose of 0.25 mg/kg body weight once every other day for four doses.
Tissue Collection and Immunohistochemistry.
At time points indicated for tissue harvest, mice were killed by isoflurane overdose. Lungs were inflated with phosphate-buffered saline (PBS) administered through the trachea. Left, superior, and postcaval lobes were isolated, fixed overnight in formalin at 4 °C, then transferred to 70% ethanol. After processing and paraffin embedding, tissue sections were cut to a thickness of 5 μm. Hematoxylin and eosin (H&E) staining was performed according to standard protocol. For immunohistochemical (IHC) staining, slides were deparaffinized and treated with Antigen Unmasking Solution (Vector Laboratories, H-3300); then, endogenous peroxidases were inactivated by incubation in 3% hydrogen peroxide (Sigma). Primary antibodies used (with dilutions) are as follows: AGO2 (Sino Biologicals, 50683-RO-36) 1:100; Ki67 (BD Biosciences, 550609) 1:500; Phospho-Erk1/2 (Cell Signaling, 4376S) 1:100; and Phospho-Akt (Cell Signaling, 4060S) 1:100 were diluted in 10% normal goat serum (NGS) (in PBS). Antibody detection was achieved with species-specific VECTSTAIN Elite ABC kits (Vector Labs) and a DAB Peroxidase Substrate kit (Vector Labs). All slides were analyzed by a board-certified pathologist.
Mouse Lung Nodule Isolation and Ex Vivo Culture.
Lung dissociation and organoid culture were performed as previously described (41, 42) with scissors. Samples were then digested for 1 h at 37 °C in a mixture of dispase (1 mg/mL) (BD Biosciences), trypsin/2 mM ethylenediaminetetraacetic acid (EDTA) (0.25% by volume) (Cellgro), and elastase (10 U/mL) (Worthington). Dissociated cells were passed through a 70-μm cell strainer and resuspended in organoid culture medium (OCM) containing the following: Dulbecco's Modified Eagle Medium (DMEM)/F12 supplemented with 1xITS (insulin, transferrin, and selenium; Invitrogen), 0.5% BPE (bovine pituitary extract; Fisher), 25 ng/mL mEGF (epidermal growth factor; PeproTech), 0.1 μg/mL CTX (cholera toxin; Sigma), 10 μM Y-27632 (ROCK inhibitor; Sigma), 100 nM retinoic acid (Cayman), and 100 units /mL penicillin–streptomycin (Gibco, Cat. No. 10378016). Aliquots of 1 × 104 cells, in 50 μL OCM, were mixed with 50 μL Growth Factor Reduced Matrigel (Corning) and plated into the center of single wells in Poly-D-Lysine Cellware 12-well plates (Corning). After 30 min in a 37 °C cell culture incubator supplemented with 5% CO2, OCM was added to each well and changed every 2 d for the duration of the experiment. Two-dimensional culture of organoid cells for in vitro transfection experiments was performed as follows: Trypsinized organoid pellets were transferred to six-well collagen-coated plates (Corning) containing OCM. Cells were allowed to adhere overnight after which they were transfected with indicated AGO2 constructs (12, 13) using Lipofectamine 3000 (Thermo Fisher Scientific). Proliferation of cultured cells was analyzed using the Incucyte live cell imaging system (Sartorius). Lysis and immunoblotting (described below) were performed 72 h after transfection.
Immunoblotting and Immunoprecipitation.
Cells were lysed in ice-cold radioimmunoprecipitation assay (RIPA) buffer (Pierce) supplemented with phosphatase inhibitor mixture. Protein concentration was the quantified using the Bio-Rad Protein Assay. Between 10 and 30 μg of total protein was separated on 4–12% Bis-Tri gradient gels (Life Technoloigies) by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to nitrocellulose membranes. Antibodies are indicated as follows: Ago2 (Sino Biologicals, 50683-RO-36) 1:1,000; GAPDH(14C10) (Cell Signaling, 3683S) 1:1,000; H3 (Cell Signaling, 9715) 1:1,000; Phospho-Erk1/2 (Cell Signaling, 4376S) 1:1,000; Erk1/2 (Cell Signaling, 9102S) 1:1,000; Phospho-Akt (Cell Signaling, 4060S) 1:1,000; and Akt (Cell Signaling, 2920S) 1:1,000. Immunoprecipitation experiments were carried out using 300 μg cell lysate. This was precleared with Protein A/G agarose beads for 1 h at 4 °C. Following incubation, lysates were treated with 5 μg of Ras antibody (Ras10 monoclonal antibody, Millipore) or 5 μg IgG isotype control overnight at 4 °C. Subsequently, Protein A/G agarose beads were used to precipitate immune complexes (2-h incubation at 4 °C). Immunoprecipitates were then washed, resuspended in loading buffer, and analyzed by immunoblot.
Transcriptome Analysis.
mRNA isolated as indicated above was quantitated on the Illumina platform using RiboErase methodology. Transcripts were quantified by pseudoalignment algorithm Kallisto (53) with mm10 as the reference genome. Estimated counts were used to create DEGList and normalized by trimmed mean of M values (TMM) (54) using the calcNormFactors of edgeR (55). Lowly expressed genes with mean transcript per million (TPM) of less than 1 were removed from downstream analysis. Differential analysis between biallelic knockout and wild type was performed on voom-transformed count data (56) using the limma (57) package. Finally, the fgsea package was used to perform gene set analysis using estimated log-fold change from differential analysis as input. Mouse genes were converted to human homologs using biomaRT (58) before evaluating enrichment of gene sets. Enrichment plots and heat maps were generated in R Studio.
MicroRNA Expression Profiles Using Quantitative PCR of Mouse miRnome Panels.
Total RNA was isolated from lung nodules using the AllPrep DNA/RNA/miRNA Universal Kit (Qiagen). Five nanograms of RNA from each sample was converted into cDNA using the miRCURY LNA Universal RT microRNA PCR Universal cDNA Synthesis Kit II. Quantitative micro RT-PCR was performed using exiLENT SYBR Green master mix with microRNA ready-to-use PCR mix, Mouse&Rat panel I, V4.M (Exiqon) on an ABI 7900HT Fast Real Time PCR system (Applied Biosystems). Data were analyzed using GenEX version 6 software.
Statistical Analysis.
Intergroup comparisons were made with Student’s t test (two-group comparisons) or one-way ANOVA (three-group comparisons). For RNA-seq, a moderated t test implemented with limma was used for group comparison and P value adjustment for multitest was made with the Benjamini–Hochberg (BH) method. For microRNA, significance was tested with a Wilcoxon signed-rank test. Statistical significance was defined as a P value <0.05 (BH adjusted P value for RNA-seq).
Supplementary Material
Acknowledgments
This work was supported by the Prostate Cancer Foundation, Prostate SPORE Grant P50 CA186786, National Cancer Institute Outstanding Investigator Award R35 CA231996, and the Early Detection Research Network U01 NIH Grant CA214170. A.M.C. is a Howard Hughes Medical Institute Investigator, an A. Alfred Taubman Scholar, and an American Cancer Society Professor.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2026104118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Siegel R. L., Miller K. D., Jemal A., Cancer statistics, 2017. CA Cancer J. Clin. 67, 7–30 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Ettinger D. S.et al.; NCCN (National Comprehensive Cancer Network) , Non-small cell lung cancer. J. Natl. Compr. Canc. Netw. 10, 1236–1271 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Schubbert S., Shannon K., Bollag G., Hyperactive Ras in developmental disorders and cancer. Nat. Rev. Cancer 7, 295–308 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Karnoub A. E., Weinberg R. A., Ras oncogenes: Split personalities. Nat. Rev. Mol. Cell Biol. 9, 517–531 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trahey M., McCormick F., A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science 238, 542–545 (1987). [DOI] [PubMed] [Google Scholar]
- 6.Shaw R. J., Cantley L. C., Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature 441, 424–430 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Garrido P., et al., Treating KRAS-mutant NSCLC: Latest evidence and clinical consequences. Ther. Adv. Med. Oncol. 9, 589–597 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox A. D., Fesik S. W., Kimmelman A. C., Luo J., Der C. J., Drugging the undruggable RAS: Mission possible? Nat. Rev. Drug Discov. 13, 828–851 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindsay C. R., Jamal-Hanjani M., Forster M., Blackhall F., KRAS: Reasons for optimism in lung cancer. Eur. J. Cancer 99, 20–27 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Janes M. R., et al., Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell 172, 578–589.e17 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Hong D. S., et al., KRASG12C inhibition with sotorasib in advanced solid tumors. N. Engl. J. Med. 383, 1207–1217 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shankar S., et al., KRAS engages AGO2 to enhance cellular transformation. Cell Rep. 14, 1448–1461 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shankar S.et al., An essential role for Argonaute 2 in EGFR-KRAS signaling in pancreatic cancer development. Nat. Commun. 11, 2817 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sasaki T., Shiohama A., Minoshima S., Shimizu N., Identification of eight members of the Argonaute family in the human genome. Genomics 82, 323–330 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Valdmanis P. N., et al., Expression determinants of mammalian Argonaute proteins in mediating gene silencing. Nucleic Acids Res. 40, 3704–3713 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutvágner G., Zamore P. D., A microRNA in a multiple-turnover RNAi enzyme complex. Science 297, 2056–2060 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Hutvagner G., Simard M. J., Argonaute proteins: Key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 9, 22–32 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Meister G., et al., Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 15, 185–197 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Cheloufi S., Dos Santos C. O., Chong M. M., Hannon G. J., A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465, 584–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frohn A., et al., Dicer-dependent and -independent Argonaute2 protein interaction networks in mammalian cells. Mol. Cell. Proteomics 11, 1442–1456 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winter J., Diederichs S., Argonaute proteins regulate microRNA stability: Increased microRNA abundance by Argonaute proteins is due to microRNA stabilization. RNA Biol. 8, 1149–1157 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Lu J., et al., MicroRNA expression profiles classify human cancers. Nature 435, 834–838 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Takamizawa J., et al., Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 64, 3753–3756 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Chirshev E., Oberg K. C., Ioffe Y. J., Unternaehrer J. J., Let-7 as biomarker, prognostic indicator, and therapy for precision medicine in cancer. Clin. Transl. Med. 8, 24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L., Yu C., Gao H., Li Y., Argonaute proteins: Potential biomarkers for human colon cancer. BMC Cancer 10, 38 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng N., Li Y., Han Z. G., Argonaute2 promotes tumor metastasis by way of up-regulating focal adhesion kinase expression in hepatocellular carcinoma. Hepatology 57, 1906–1918 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Vaksman O., Hetland T. E., Trope’ C. G., Reich R., Davidson B., Argonaute, Dicer, and Drosha are up-regulated along tumor progression in serous ovarian carcinoma. Hum. Pathol. 43, 2062–2069 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Shen J., et al., EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature 497, 383–387 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J., et al., Up-regulation of Ago2 expression in gastric carcinoma. Med. Oncol. 30, 628 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Cerami E., et al., The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DuPage M., Dooley A. L., Jacks T., Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat. Protoc. 4, 1064–1072 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheridan C., Downward J., Overview of KRAS-driven genetically engineered mouse models of non-small cell lung cancer. Curr. Protoc. Pharmacol. 70, 14 35 1–14 35 16 (2015). [DOI] [PubMed] [Google Scholar]
- 33.O’Carroll D., et al., A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev. 21, 1999–2004 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rawlins E. L., et al., The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell 4, 525–534 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ji H., et al., K-ras activation generates an inflammatory response in lung tumors. Oncogene 25, 2105–2112 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Xu X., et al., The cell of origin and subtype of K-Ras-induced lung tumors are modified by Notch and Sox2. Genes Dev. 28, 1929–1939 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sweet-Cordero A., et al., An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat. Genet. 37, 48–55 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Chen X., Du J., Jiang R., Li L., MicroRNA-214 inhibits the proliferation and invasion of lung carcinoma cells by targeting JAK1. Am. J. Transl. Res. 10, 1164–1171 (2018). [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao X., et al., MicroRNA-214 governs lung cancer growth and metastasis by targeting carboxypeptidase-D. DNA Cell Biol. 35, 715–721 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Li Y., Zhao L., Qi Y., Yang X., MicroRNA-214 upregulates HIF-1α and VEGF by targeting ING4 in lung cancer cells. Mol. Med. Rep. 19, 4935–4945 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Barkauskas C. E., et al., Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 123, 3025–3036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng T., et al., Hedgehog actively maintains adult lung quiescence and regulates repair and regeneration. Nature 526, 578–582 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dearden S., Stevens J., Wu Y. L., Blowers D., Mutation incidence and coincidence in non small-cell lung cancer: meta-analyses by ethnicity and histology (mutMap). Ann. Oncol. 24, 2371–2376 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uras I. Z., Moll H. P., Casanova E., Targeting KRAS mutant non-small-cell lung cancer: Past, present and future. Int. J. Mol. Sci. 21, 4325 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skoulidis F., et al., Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 5, 860–877 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jackson E. L., et al., Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 15, 3243–3248 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blasco R. B., et al., c-Raf, but not B-Raf, is essential for development of K-Ras oncogene-driven non-small cell lung carcinoma. Cancer Cell 19, 652–663 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Engelman J. A., et al., Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat. Med. 14, 1351–1356 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trejo C. L., Juan J., Vicent S., Sweet-Cordero A., McMahon M., MEK1/2 inhibition elicits regression of autochthonous lung tumors induced by KRASG12D or BRAFV600E. Cancer Res. 72, 3048–3059 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jänne P. A., et al., Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: A randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol. 14, 38–47 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Cicchini M., et al., Context-dependent effects of amplified MAPK signaling during lung adenocarcinoma initiation and progression. Cell Rep. 18, 1958–1969 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marino S., Vooijs M., van Der Gulden H., Jonkers J., Berns A., Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 14, 994–1004 (2000). [PMC free article] [PubMed] [Google Scholar]
- 53.Bray N. L., Pimentel H., Melsted P., Pachter L., Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Robinson M. D., Oshlack A., A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Law C. W., Chen Y., Shi W., Smyth G. K., voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ritchie M. E., et al., Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Durinck S., Spellman P. T., Birney E., Huber W., Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4, 1184–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.