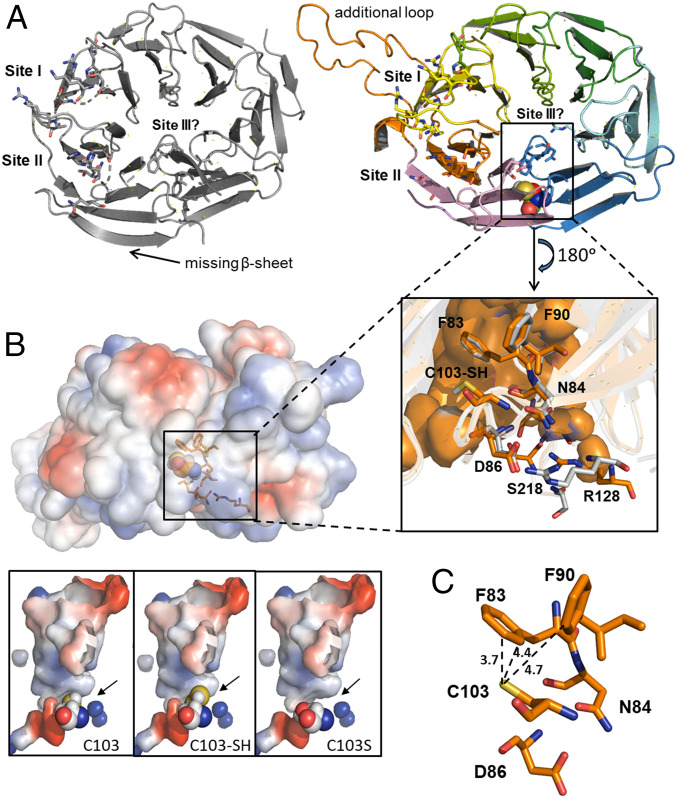
Fig. 6.
Predicted structure of AtATG18a. (A) Crystal structure of human WIPI3 (Protein Data Bank identification: 6KRL; Left) and the predicted structural model of Arabidopsis ATG18a (Right). The proposed lipid-binding sites and the cavity site of C103 are highlighted. (Inset) Residues of AtATG18a surrounding the active C103 site (F83, N84, D86, and F90) and participating in binding site III (R128 and S218). The equivalent residues in HsWIPI3 are in gray. (B) Representation of surface electrostatic potential distribution in AtATG18a structural model and zoomed into the putative conformation of the active site, showing with spheres the position of C103, the persulfidation C103-SH, and the mutation C103S. Positively and negatively charged regions are depicted in blue and red, respectively. (C) Zoom into the putative conformation of the active site showing distance (Å) between the catalytic residue C103 and F83 and F90 in AtATG18a.