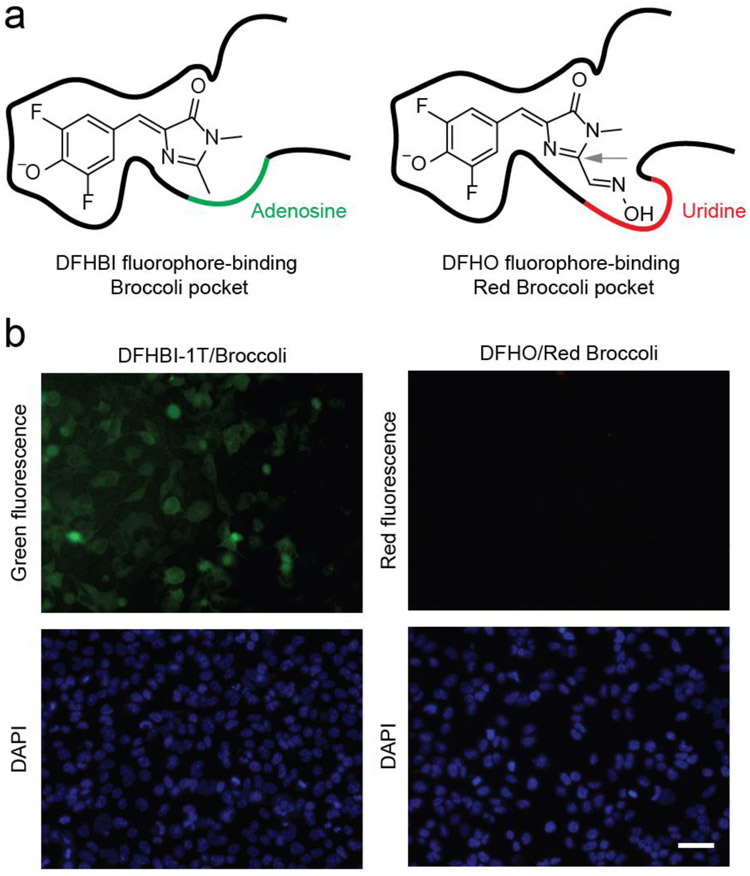
Figure 1.
Red Broccoli cannot be detected in living cells using DFHO. (a) The fluorophore-binding pocket of Red Broccoli contains a uridine (red curve), while the same position is adenosine (green curve) in Broccoli. In the left panel, Broccoli is shown with bound DFHBI, a mimic of the fluorophore naturally found in green fluorescent proteins.(10) In the right panel, Red Broccoli is schematized with bound DFHO, a mimic of the fluorophore naturally found in DsRed red fluorescent proteins.(31) The C2 position in DFHO (indicated with a gray arrow) contains an additional N-oxime group compared to that in DFHBI that confers red-shifted emission.(31) Additionally, the uridine (indicated with a red curve) in Red Broccoli appears to interact with the N-oxime.(32) (b) Red Broccoli is not readily detected in mammalian cells incubated with DFHO when cells are imaged using fluorescence microscopy. HEK293T cells expressing circular Broccoli RNA were incubated with 10 μM DFHBI-1T (left), resulting in readily detectable green fluorescent cells. Cells expressing circular Red Broccoli RNA in the presence of 10 μM DFHO (right) did not show evidence for readily detectable cellular fluorescence using these specific imaging conditions, microscope, and camera. Images were acquired using a 40× objective with a TRITC red filter cube for Red Broccoli fluorescence, a FITC green filter cube for Broccoli fluorescence, and a DAPI filter cube for Hoechst 33342 staining. Exposure times: 100 ms for the TRITC filter, 100 ms for the FITC filter, and 50 ms for the DAPI filter. Scale bar, 50 μm.