Figure 3. Myeloid cell-targeted ADAM17-deficiency suppresses pro- and anti-inflammatory responses in macrophages from APOA1Tg; Ldlr−/− mice.
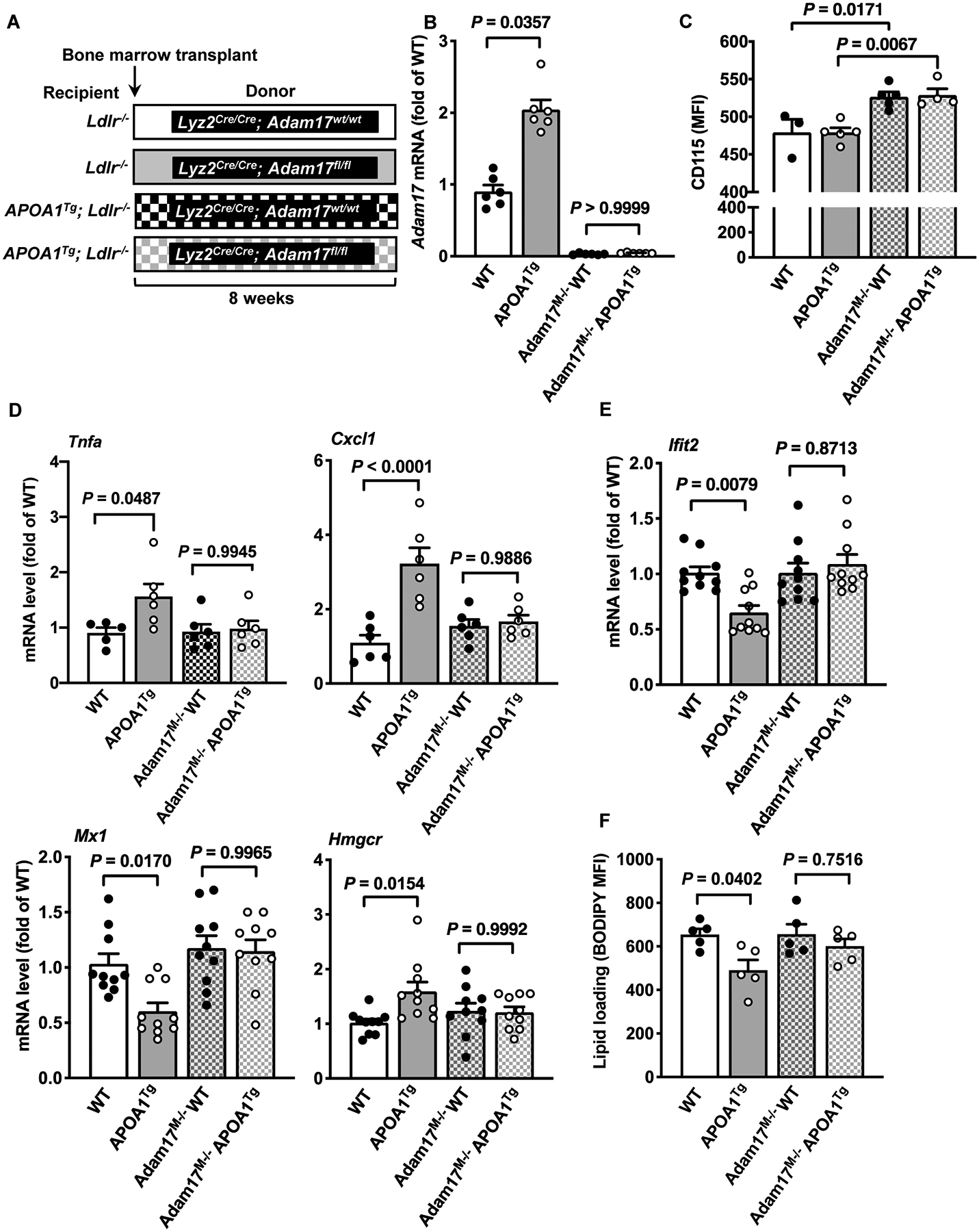
A. Bone marrow was harvested from myeloid cell-targeted ADAM17-deficient mice or WT C57BL/6J littermates and transplanted into lethally irradiated (10 Gy) male Ldlr−/− or APOA1Tg; Ldlr−/− recipient littermates. After 8 weeks of recovery, peritoneal cells from Ldlr−/− and APOA1Tg; Ldlr−/− recipient mice were collected 4 days after thioglycolate injection, macrophages were purified from other cell types using a macrophage isolation kit followed by a 1 hr adhesion purification, and RNA was harvested for further analysis. B. Adam17 mRNA levels analyzed in macrophages from Ldlr−/− and APOA1Tg; Ldlr−/− recipient mice (n=5–6). C. Blood leukocytes from Ldlr−/− and APOA1Tg; Ldlr−/− recipient mice stained with an anti-CD115 antibody and analyzed using flow cytometry (n=3–5). D. Macrophages from Ldlr−/− and APOA1Tg; Ldlr−/− mice stimulated with LPS (10 ng/ml, 4 hr). At the end of the treatment, the cells were used to analyze Tnfa mRNA and Cxcl1 (n=5). E. Macrophages from Ldlr−/− and APOA1Tg; Ldlr−/− mice were stimulated with IFNβ (1 ng/ml, 4 hr). At the end of the treatment, the cells were used to analyze gene expression of Ifit2, Mx1 and Hmgcr (n=10). F. Macrophages from Ldlr−/− and APOA1Tg; Ldlr−/− recipient mice stained with BODIPY® 493/503 and analyzed using flow cytometry for neutral lipids (n=5). Data are shown as mean ± SEM. Tests for normality (Shapiro-Wilk) and equal variance (Brown-Forsythe) were performed for each of the data sets. P values were determined accordingly by one-way ANOVA followed by Tukey’s multiple comparison tests (C, D, E, F) or Brown-Forsythe ANOVA followed by Dunnett’s multiple comparisons tests (B). One significant outlier (Grubb’s test) was excluded in the WT group in D (Tnfa).
