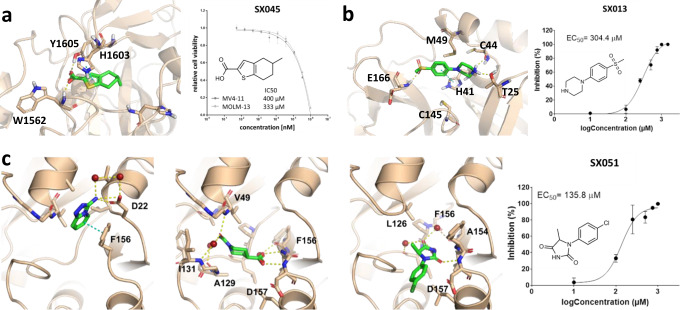
Fig. 4. Binding poses and cellular activities of SpotXplorer0 hits against challenging and current targets.
a Predicted binding pose of the fragment SX045 (green) in the binding pocket of the SETD2 histone methyltransferase. The fragment decreases the viability of MV4-11 and MOLM-13 leukemia cells in a dose-dependent manner (see Supplementary Information, section 8), with IC50 values of 400 µM and 333 µM, respectively. b X-ray structure of the fragment SX013 in complex with the main protease (3CLPro) of the SARS-CoV-2 virus (PDB entry 5RHD). The fragment inhibits the SARS-COV-2-induced mortality of Vero E6 cells with an EC50 of 304 µM. c X-ray structures of fragments SX005, SX048 and SX051 (left to right) in complex with the NSP3 macrodomain of the SARS-CoV-2 virus (PDB entries 5S4G, 5S4H, and 5S4I). The fragments show antiviral activities with EC50 values in the high micromolar range in the cellular assay. (In the IC50 and EC50 plots, data are presented as mean values +/− SD, calculated from n = 3 biologically independent samples.) Source data are provided in the Source Data file.