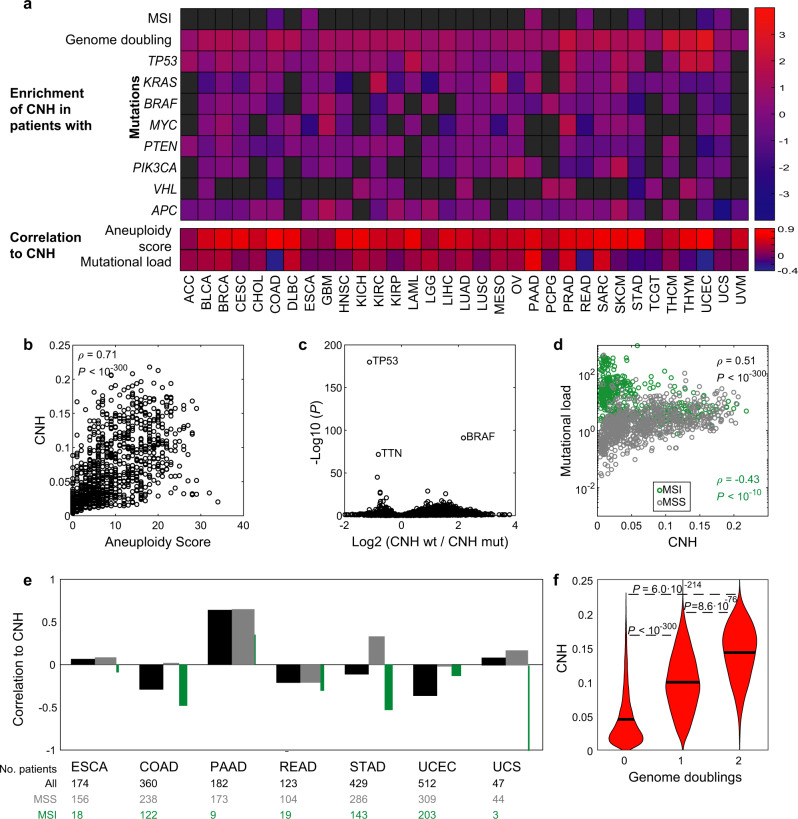
Fig. 3. Genetic characterization of CNH.
a Genomic associations per type for all 33 cancer types in TCGA. Upper panel: enrichment of copy number heterogeneity (CNH) in microsatellite-instable (MSI), genome-doubled or mutated cancers versus non-MSI, non-genome-doubled and non-mutated cancers, respectively. The enrichment of CNH is defined as the log2 of the ratio between group medians with and without event. Lower panel: Spearman’s rank correlation of CNH to aneuploidy and mutational load per cancer type. b Pan-cancer Spearman’s rank correlation between CNH and the aneuploidy score. c Identification of mutated genes related to CNH in a pan-cancer setting. Malignancies are grouped as wild type or mutated for each gene. The relative difference in median CNH of these groups (horizontal axis) and the corresponding significance calculated by the Wilcoxon rank-sum test (vertical axis) are shown. d Spearman’s rank correlation between CNH and mutational load in MSI tumours (green) and non-MSI cancers (grey). e Spearman’s rank correlation between CNH and mutational load per cancer type for all tumours (black), non-MSI tumours (grey) and MSI tumours (green). The relative width of the grey and green bars reflects the ratio of MSI/non-MSI tumours in each type. f Malignancies that have undergone genome doubling have a higher CNH. Groups are compared by the Wilcoxon rank-sum test. Source data are provided as a Source Data file.