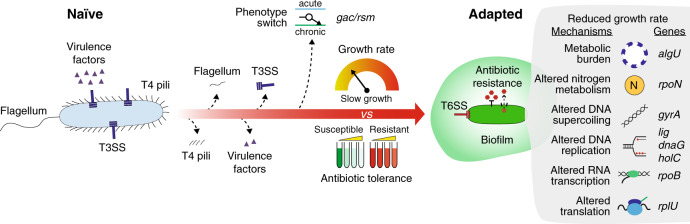
Fig. 6. Trajectory of adaptation of Pseudomonas aeruginosa to the cystic fibrosis (CF) environment.
The cartoon represents the trajectories of evolution from naïve to adapted strain occurring in the CF patients. Several cellular components such as the flagellum, the type 4 pili (T4 pili), the type 3 secretion system (T3SS) and numerous virulence factors are lost during within-patient evolution due to accumulation of mutations. The Gac/Rsm regulatory system coordinates the switch between acute and chronic phenotype through changes of gene expression profiles. Meanwhile, a reduction of the growth rate occurs due to: (1) metabolic burden caused by algU regulator overexpression; (2) altered nitrogen metabolism caused by mutations in rpoN and caused by changes in the RpoN regulon transcriptional profiles; (3) altered DNA supercoiling caused by gyrA gene mutations; (4) altered DNA replication caused by mutations on important component of the DNA polymerase (lig, dnaG, holC); (5) altered transcription caused by rpoB gene mutations; and (6) altered translation caused by mutations in ribosomal proteins genes. Such reduction in growth rate determines an increased tolerance to antibiotics which finally helps P. aeruginosa to persist in the CF lungs.