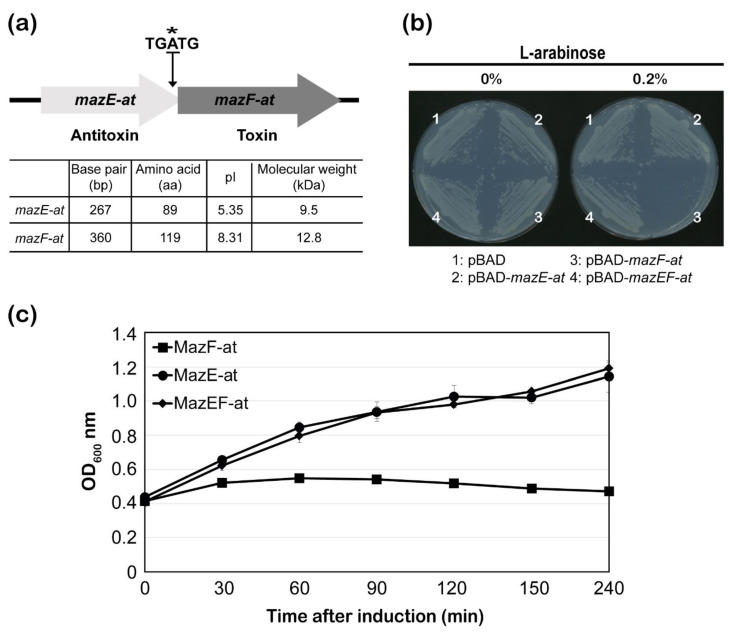
Figure 1.
The characterization of the MazEF-at TA system in Agrobacterium tumefaciens. (a) Gene map of the mazEF-a. The asterisk indicates the nucleotide where the orf of the two genes overlap. The table includes the gene length, number of amino acids, theoretical pI, and molecular weight of MazE-at and MazF-at; (b) Toxicity of MazF-at on plates. The cells of Escherichia coli BW25113 harboring pBAD24-mazE-at (2), -mazF-at toxin (3), -mazEF-at (4) or pBAD24 only (1) were incubated on the M9 plate with or without 0.2% L-arabinose; (c) Effect of MazF-at on cell growth induction. The culture of E. coli BW25113 harboring pBAD24-mazE-at (black circle), -mazF-at (black square), or –mazEF-at (black rhombus) was grown until the OD 600nm (optical density) of the culture reached ~0.4, followed by adding 0.2% L-arabinose at 0 min, and bacterial growth was measured for 6 h at OD600 nm. Error bars represent the standard error of the mean (SEM).